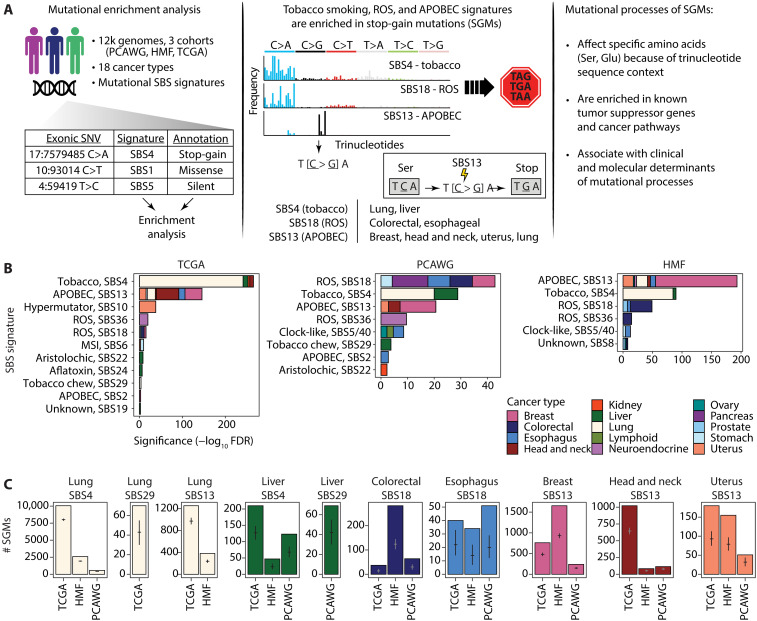
Fig. 1. Protein-coding impact of mutational signatures in cancer genomes and associations with SGMs.
(A) Overview of the study. Left: The associations of protein-coding impact of somatic SNVs and mutational signatures of SBSs were studied using enrichment analysis in >12,000 cancer genomes. Middle: SGMs were enriched in the SBS signatures of tobacco smoking, APOBEC, and ROS. These are explained by the trinucleotide preferences of the mutational processes that affect certain amino acid codons and convert these to stop codons. Right: Mutational signatures of SGMs were further studied in the context of driver genes and pathways, as well as the clinical and molecular correlates of the mutational processes. (B) Enrichments of mutational signatures in SGMs in multiple cancer types and in the three cancer genomics datasets (TCGA, HMF, and PCAWG) (FDR < 0.01). Bar plots show the cumulative significance of enriched SBS signatures in SGMs in various types of cancer. Tobacco smoking, APOBEC activity, and ROS exposure are the major mutational processes that contribute SGMs in multiple cancer types. (C) Observed and expected counts of SGMs derived from the most significant mutational processes. Mean expected mutation counts with 95% confidence intervals (CI) from binomial sampling are shown on the bars.