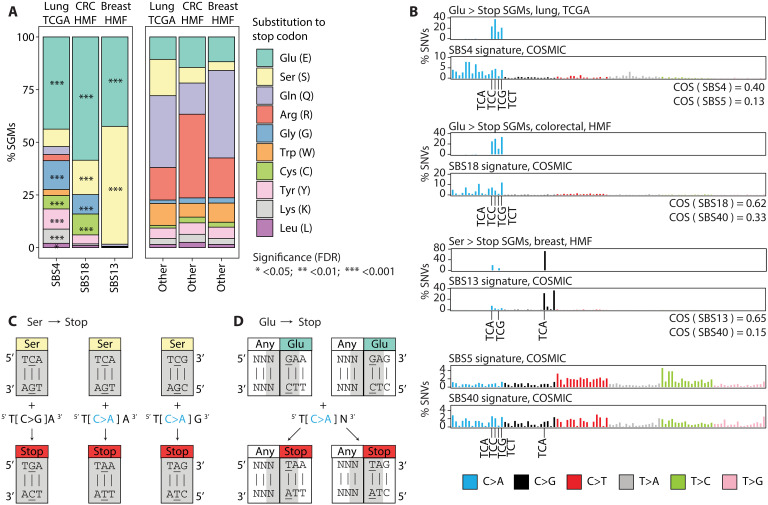
Fig. 2. SBS signatures induce protein-truncating mutations by targeting the genetic code.
(A) SGMs of tobacco smoking, APOBEC, and ROS signatures are enriched in specific amino acids. Bar plots show the proportions of amino acids affected by SGMs. Left: SGMs of the three major signatures: SBS4 in lung cancer, SBS18 in colorectal cancer, and SBS13 in breast cancer. Right: Control SGMs of all other mutational signatures in these cancer types. Enrichment of signature-associated SGMs relative to controls are shown as asterisks (Fisher’s exact tests). (B) Trinucleotide profiles of SGMs of glutamic acid and serine residues (Glu>Stop, Ser>Stop) and the reference COSMIC signatures for SBS4, SBS18, and SBS13. As controls, the profiles of the most frequent SBS signatures are shown (SBS5/40; two plots at the bottom). Cosine similarity (COS) scores comparing the SGM signatures and the COSMIC reference signatures are shown. The trinucleotide substitutions from (C) and (D) are highlighted on the x axis. (C and D) SBS signatures interact with the genetic code to induce stop codons. The codons encoding serines and glutamic acids are shown as rectangles. The trinucleotides inducing stop codons upon mutation are shown in gray. Base mutations are underlined. (C) Serine substitutions to stop codons. C>G and C>A transversions in SBS13 and SBS18 induce stop codons by substituting the middle nucleotides of serine codons (yellow). (D) Glutamic acid substitutions to stop codons. C>A transversions in SBS4, SBS13, and SBS18 induce stop codons by affecting two consecutive codons. Because SBS signatures are represented with pyrimidines as reference nucleotides, this schematic shows the reverse complements of the trinucleotides in which glutamic acids are changed to stop codons. Here, the reverse-complementary trinucleotide transversions characteristic of the tobacco smoking signature SBS4 substitute the first nucleotide in the glutamic acid codon (teal).