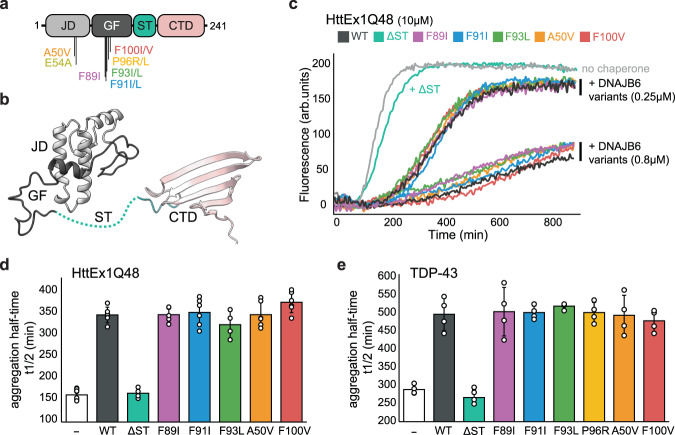
Fig. 1. LGMDD1 DNAJB6 mutants efficiently suppress protein aggregation.
a Domain organization of DNAJB6 showing the positions of the LGMDD1-associated mutations. All mutations identified to date are located in the N-terminal section of the protein, primarily in the GF-rich region. JD is colored light gray, the GF region dark gray, the ST-rich region turquoise, and the C-terminal domain pink. b NMR structure of DNAJB6 lacking the ST oligomerization region (PDB 6U3R19), with domains colored as in (a). c Aggregation of 10 µM HTTEx1-Q48 alone (light gray) and in the presence of two concentrations (0.25 and 0.8 µM) of WT DNAJB6 (dark gray) or LGMDD1 disease mutants (colored). The mutants are as efficient as the WT chaperone in preventing aggregation. d The effect of 0.8 µM DNAJB6 WT and disease mutants on HTTEx1-Q48 (10 µM) aggregation half times. Data represents mean values ± s.d (n = 6 independent experiments for control, DNAJB6 WT, F91L, A50V, and F100V variants; n = 5 for F89I; n = 4 for DNAJB6 ΔST and F93L variants). e The effect of 5 µM DNAJB6 WT and disease mutants on 10 µM TDP-43 aggregation half times. Data represents mean values ± s.d (n = 4 independent experiments).