Figure 2.
Three-level integration data
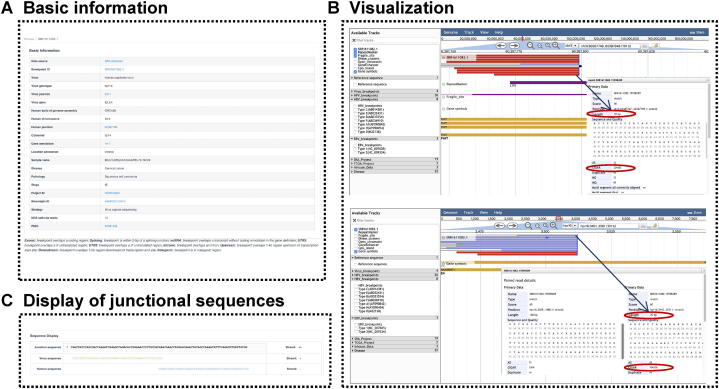
A. Basic information. B. Manual visualization of supporting reads for NGS-derived computational breakpoints in VIS Browser. This function was built based on the JBrowse genome browser, which was equipped with the human genome (GRCh38), related multi-omics tracks, and all needed virus genomes. SRR1611082.1 (Breakpoint ID), one HPV16 integration site in FHIT is given as an example. The top is the mapping view of soft-clip reads in FHIT, below is that in the HPV16 genome within the 101-bp window around the integration sites. The supported read SRR1611082.15598289 (Read ID) is 100 bp in length with 32 bp mapped to the human genome and 70 bp to the virus genome (2-bp MH shared by human and virus). C. Display of junctional sequences. Chr, chromosome.