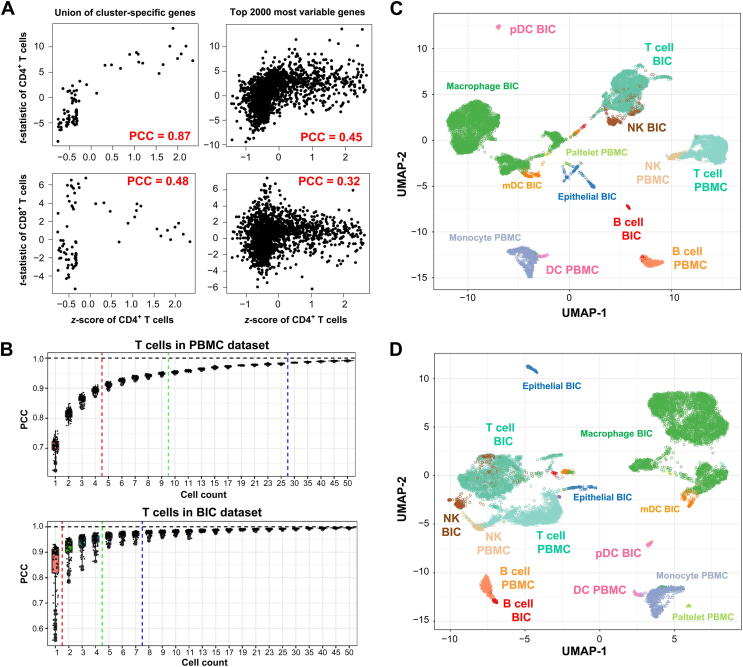
Figure 6.
deCS pipeline reduces the collinearity and impact of sequencing depth difference
A. Correlation comparison by using the union of cell cluster-specific genes and top 2000 most variable genes between query (CD4+ T) cells and reference on CD4+ T (positive; upper panels) or CD8+ T cells (negative; lower panels). B. Rarefaction curve of T cells in PBMC and BIC datasets. X-axis indicates the sized pools (n from 1 to 50). Y-axis indicates the inter-correlation between the gene expression of pooled cells and pseudo-bulk level (cluster averaged). The red, green, and blue lines indicate the number of single cells that hypothetically enables the averaged expression levels of pooled cells approximate to bulk populations with PCC > 0.90, PCC > 0.95, and PCC > 0.975, respectively. UMAP integrative analysis of PBMC and BIC datasets by natural-log-normalized expression matrix (C) or further z-score scaled (D). Each circle or triangle represents a single cell derived from BIC or PBMC dataset. PBMC, peripheral blood mononuclear cell; UMAP, uniform manifold approximation and projection.