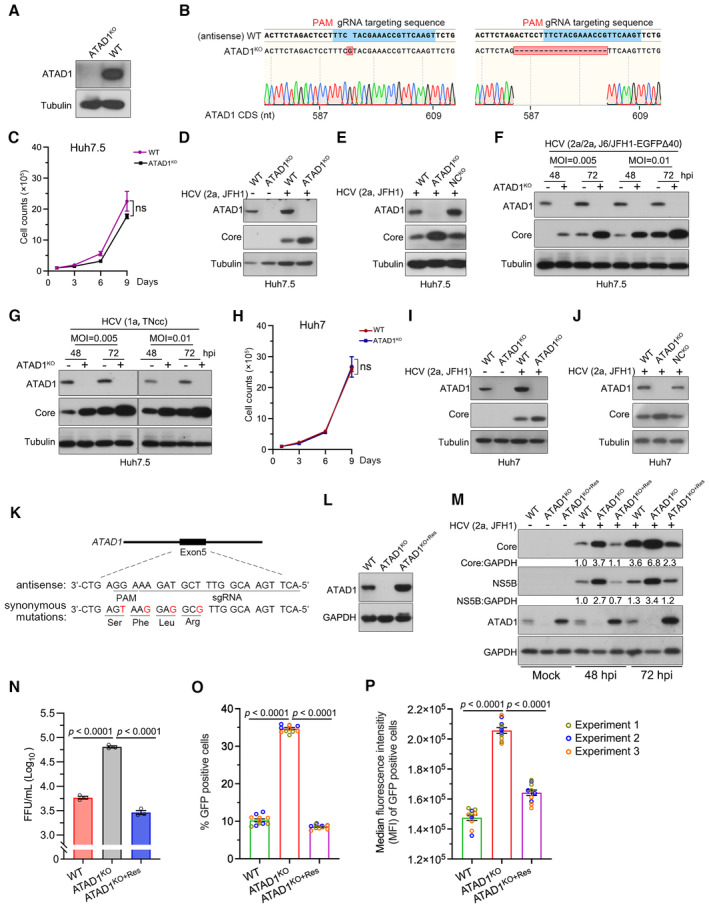
Figure 1. ATAD1 suppressed HCV infection in cultured hepatoma cells.
-
AGeneration of ATAD1KO cells. WT and ATAD1KO Huh7.5 cells were harvested and analyzed by western blotting with an anti‐ATAD1 antibody.
-
BSequencing analysis of ATAD1KO Huh7.5 cells. The genome of ATAD1KO Huh7.5 cells was extracted, and a region spanning sgATAD1 RNA‐targeting sequence was amplified by PCR. The PCR products were cloned for Sanger sequencing analysis. Seventeen clones of PCR products were sequenced, and 1‐nt insertion (n = 6) and 19‐nt deletion (n = 11) were identified in the clonal analysis.
-
CGrowth kinetics of WT and ATAD1KO Huh7.5 cells. The 1 × 105 cells were seeded and the cell counts were determined every 2 days. Data are from three independent experiments (n = 3).
-
DHCV infection in WT and ATAD1KO Huh7.5 cells. The cells were infected with HCV genotype 2a JFH1, and the cell lysates were collected for analysis by western blotting 48 h post‐infection (hpi). “+” and “−” indicate with HCV and without HCV infection, respectively.
-
EHCV infection in WT, ATAD1KO, and NCKO Huh7.5 cells. The cells were infected with JFH1 for 48 h. The NCKO was generated by transduction of lentivirus expressing irrelevant sgRNA‐targeting GFP.
-
F, GInfections of HCV genotypes 1a and 2a. WT and ATAD1KO Huh7.5 cells were infected with J6/JFH1‐EGFPΔ40 (F) and TNcc (G) at different multiplicities of infection (MOI) for 48 and 72 h.
-
HGrowth kinetics of WT and ATAD1KO Huh7 cells. The 1 × 105 cells were plated and the cell number was counted every 2 days (n = 3).
-
I, JHCV infection of WT, ATAD1KO, or NCKO Huh7 cells. The cells were infected with JFH1 for 48 h. In panels (D–G, I, and J), the cells were harvested and subjected to western blotting with anti‐ATAD1 and anti‐Core antibodies.
-
K, LThe synonymous mutations in ATAD1 complementation. The sequence of ATAD1 with four silent mutations (Mut4) was used to generate the ATAD1KO+Res Huh7.5 stable cell line (K), and the complement expression of ATAD1 in the ATAD1KO cells (ATAD1KO+Res cells) (L).
-
MHCV infection in WT, ATAD1KO, and ATAD1KO+Res Huh7.5 cells. The cells were infected with HCV 2a JFH1 (MOI = 0.015) and then the cells were collected at 48 and 72 hpi for analysis. The levels of Core and NS5B were determined by western blotting.
-
NThe infectivity titers of HCV. The FFU of supernatant virus at 48 hpi was determined from the infected culture supernatant shown in Fig EV2A (n = 3).
-
O, PThe percentage of HCV‐infected cells and the intensity of HCV protein in infected cells. WT, ATAD1KO, and ATAD1KO+Res Huh7.5 cells were infected with J6/JFH1‐EGFPΔ40 at same MOI for 72 h. Flow cytometry analysis was performed to determine the percentage of GFP‐positive cells (O) as well as the median fluorescence intensity (MFI) in HCV‐infected cells (P), which reflects the amount of HCV protein present. Three independent experiments are presented in different colors.
Data information: In panels (C, H, and N–P), data are presented as mean ± SEM from three independent experiments. The statistical significance was analyzed using unpaired two‐tailed Student's t‐test in panels (C and H). One‐way ANOVA with Dunnett's post‐hoc test was used in panels (N–P) (compared to ATAD1KO group). ns, not significant (P > 0.05).
Source data are available online for this figure.
