Figure 5. ATAD1 specifically induced the degradation of mitochondria‐localized NS5B through proteasome pathway.
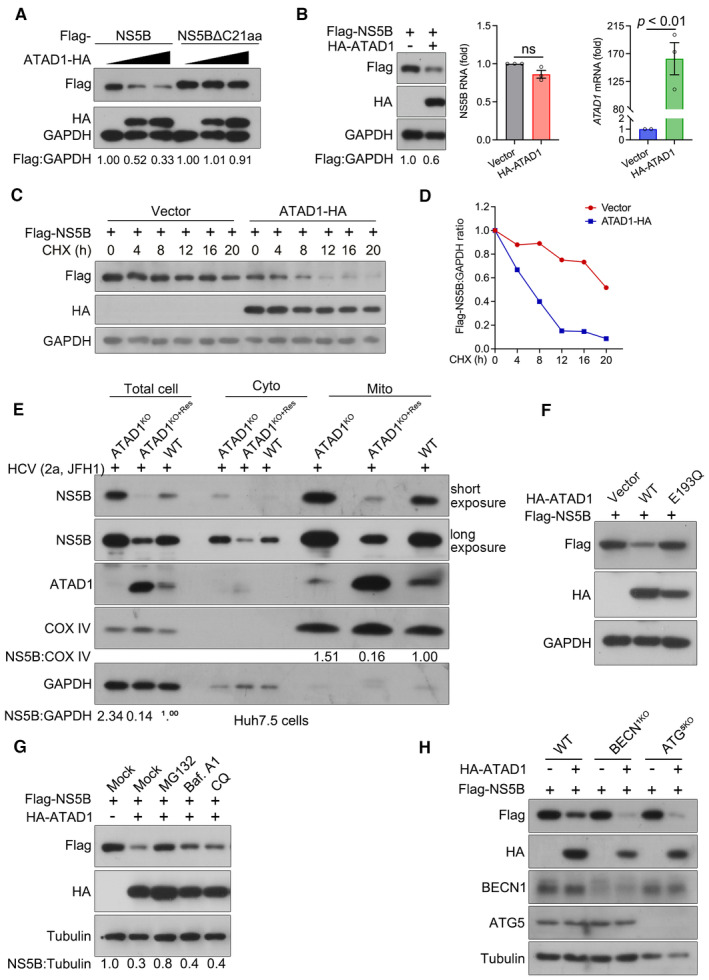
- ATAD1 mediated the degradation of NS5B. 293T cells were transfected with plasmids expressing either Flag‐NS5B or Flag‐NS5BΔC21aa, along with increasing amount of ATAD1‐HA plasmid (0, 500, and 1,500 ng). To ensure equal total plasmid amount in transfection, vector‐HA plasmid was added as a supplement. The cells were harvested at 24 h after transfection and analyzed by western blotting with anti‐Flag and anti‐HA antibodies. It is noteworthy that the mobility of full‐length Flag‐NS5B and Flag‐NS5BΔC21aa on SDS‐PAGE appears to be comparable, and was not affected by the Flag‐tag fused at either N‐ or C‐terminus (Appendix Fig S3). This observation may be attributed to the fact that the alteration in molecular weight upon removal of TMD is associated with changes in hydrophobicity, as transmembrane proteins typically possess a hydrophobic TMD while their extracellular structures are usually hydrophilic. Additionally, deletion of C21aa may have altered its interaction with SDS, resulting in changes in gel mobility.
- ATAD1 reduced the expression of NS5B protein. 293T cells were co‐transfected with Flag‐NS5B and HA‐ATAD1 or HA vector for 24 h. The cells were harvested, and then the protein and mRNA levels of Flag‐NS5B were analyzed by western blotting and qRT–PCR, respectively. The HA‐ATAD1 mRNA was also analyzed. Data are presented as mean ± SEM from three independent experiments. The statistical analysis was determined by unpaired two‐tailed Student's t‐test. ns, not significant (P > 0.05).
- CHX chase assay for NS5B protein. 293T cells were co‐transfected with Flag‐NS5B (500 ng) and ATAD1‐HA (900 ng) or vector‐HA (900 ng) for 24 h, and then the cells were treated with CHX (100 μg/ml) for indicated time duration. The cells were harvested and analyzed by western blotting with anti‐Flag and anti‐HA antibodies.
- The ratio of Flag:GAPDH in panel (C) was analyzed by gel band intensities using ImageJ software. The relative of Flag‐NS5B protein at 0 h was normalized to 1.
- Mitochondrial isolation assay. The same number of WT, ATAD1KO, and ATAD1KO+Res Huh7.5 cells were infected with JFH1 for 48 h. The cells were harvested and isolated for mitochondria using the Cell Mitochondria Isolation Kit. Equal amounts of protein (10 μg) were analyzed by western blotting with anti‐NS5B and anti‐ATAD1 antibodies. COX IV was detected as the mitochondria loading control, and GAPDH was detected as the total cell control.
- Hydrolysis activity of ATAD1 is required for the degradation of NS5B. 293T cells were transfected with plasmids expressing Flag‐NS5B and HA‐vector, Flag‐NS5B and HA‐ATAD1, or Flag‐NS5B and HA‐ATAD1‐E193Q for 24 h. The cells were harvested and lysed for western blotting with anti‐Flag and anti‐HA antibodies.
- Proteasome inhibitor prevents NS5B from degradation. 293T cells were transfected with the plasmids expressing Flag‐NS5B and HA‐ATAD1, or Flag‐NS5B and HA‐vector (mock) for 24 h, and then the cells were treated with inhibitors MG132 (10 μM), bafilomycin A1 (Baf. A1; 0.8 μM), or chloroquine (CQ; 50 μM) for 6 h and harvested for western blotting using anti‐Flag and anti‐HA antibodies.
- Degradation of NS5B independent of autophagic pathway. WT, BECN1KO, and ATG5KO 293T cells were transfected with the plasmids expressing Flag‐NS5B, together with HA‐ATAD1 or HA‐vector for 24 h. The cells were harvested and analyzed by western blotting with antibodies as indicated.
Source data are available online for this figure.
