Fig. 1. Lesional bacterial burden is associated with enhanced pro-inflammatory gene expression and delayed healing time.
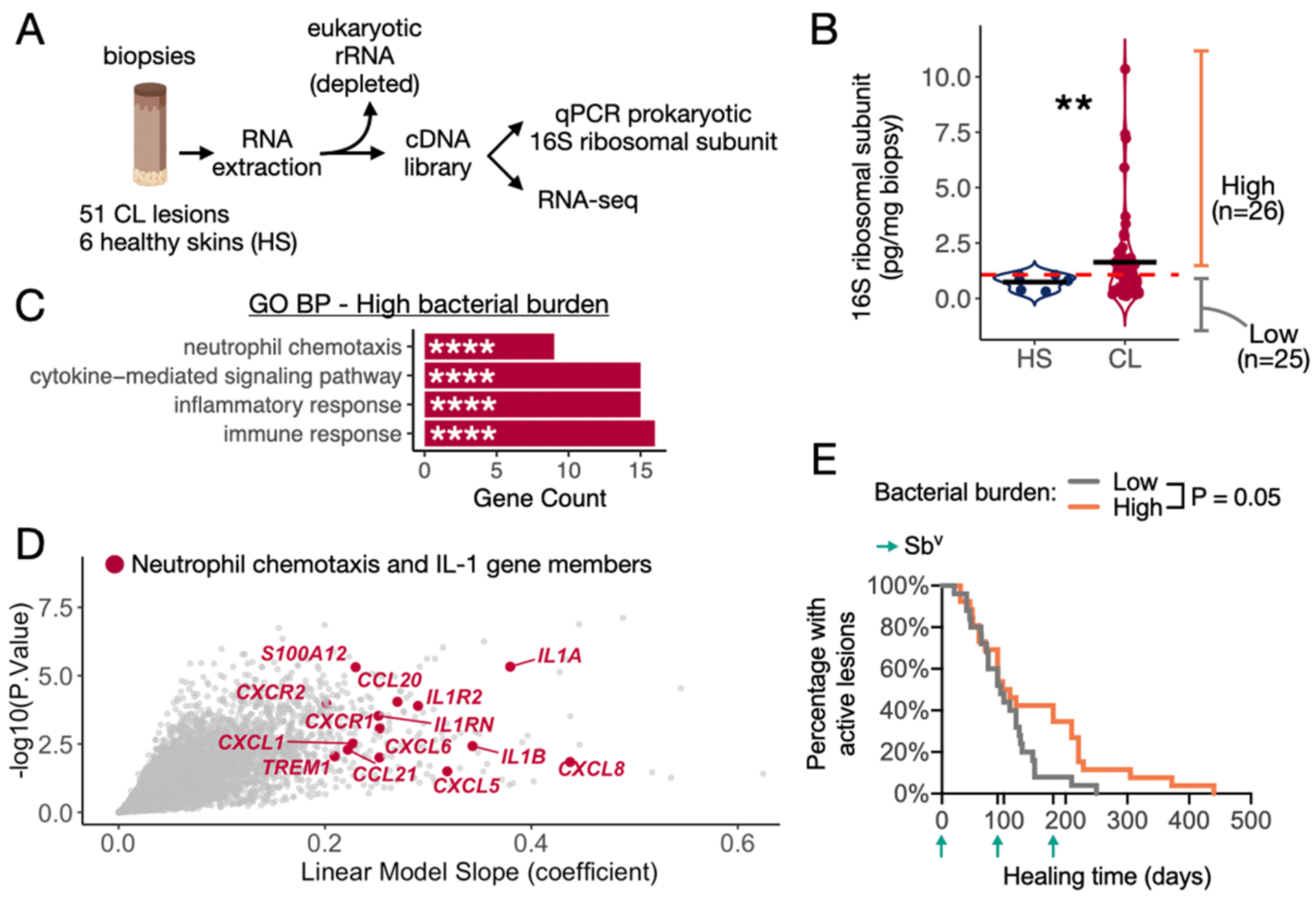
A) Schematics explaining the use of cDNA libraries from punch biopsies to perform RNA-seq and qPCR targeting the prokaryotic 16S rRNA gene to quantify total bacterial burden. Extracted RNA was depleted of eukaryotic rRNA prior to cDNA conversion. B) Bacterial burden quantified by qPCR in biopsies collected from CL lesions and healthy skin (HS) control samples not infected by L. braziliensis. The mean for each group is represented, and a T-test was used to calculate the statistical significance **P<0.01. C) GO BP analysis on the list of DEGs positively correlated with the continuous variable of bacterial burden (threshold of coefficient slope >0.2 and P<0.01). D) DEGs highlights the genes annotated as Neutrophil chemotaxis and genes encoding for members of the IL-1 signaling. E) Survival curve for lesion healing time comparing lesions with high and low bacterial burden. The threshold for dividing high and low bacterial burden lesions was based on the qPCR levels detected in HS biopsies. Log-rank (Mantel-Cox) test was used to calculate the statistical significance. GO BP, Gene Ontology with Biological Processes; ES, enrichment score. DEGs, differentially expressed genes.
