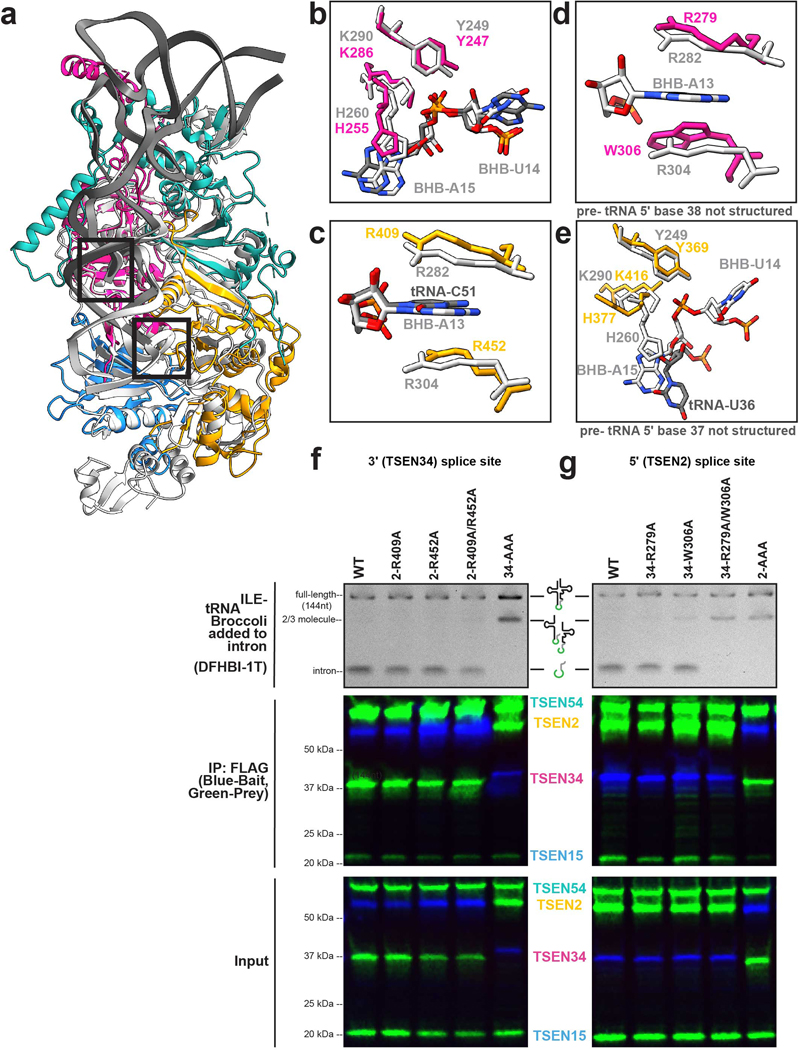
Extended Data Fig. 7 |. Structural comparison of the TSEN and EndA active site residues.
a. Overlay of TSEN + tRNA (colored as before) and EndA + BHB (grey, PDBID:2GJW) showing overall similar RNA binding. b. Overlay of the active site residues for the 3’ splice site and the c. cation-π residues from TSEN2 shown. d. Overlay of the cation-π residues from TSEN34 and the e. TSEN2 active site residues for the 5’ splice site. f. tRNA cleavage assays of a broccoli RNA -aptamer containing pre-tRNA-ILE using samples immunoprecipitated via the mutants of the 3’ splice site (pulled down by a FLAG tag on TSEN2, except for the 34 mutant) or g. 5’ splice site (pulled down by a FLAG tag on TSEN34, except for TSEN2 active site mutant). TSEN proteins that weren’t the pull-down target had a MYC-tag. The data shown is one representative experiment of three biological replicates.