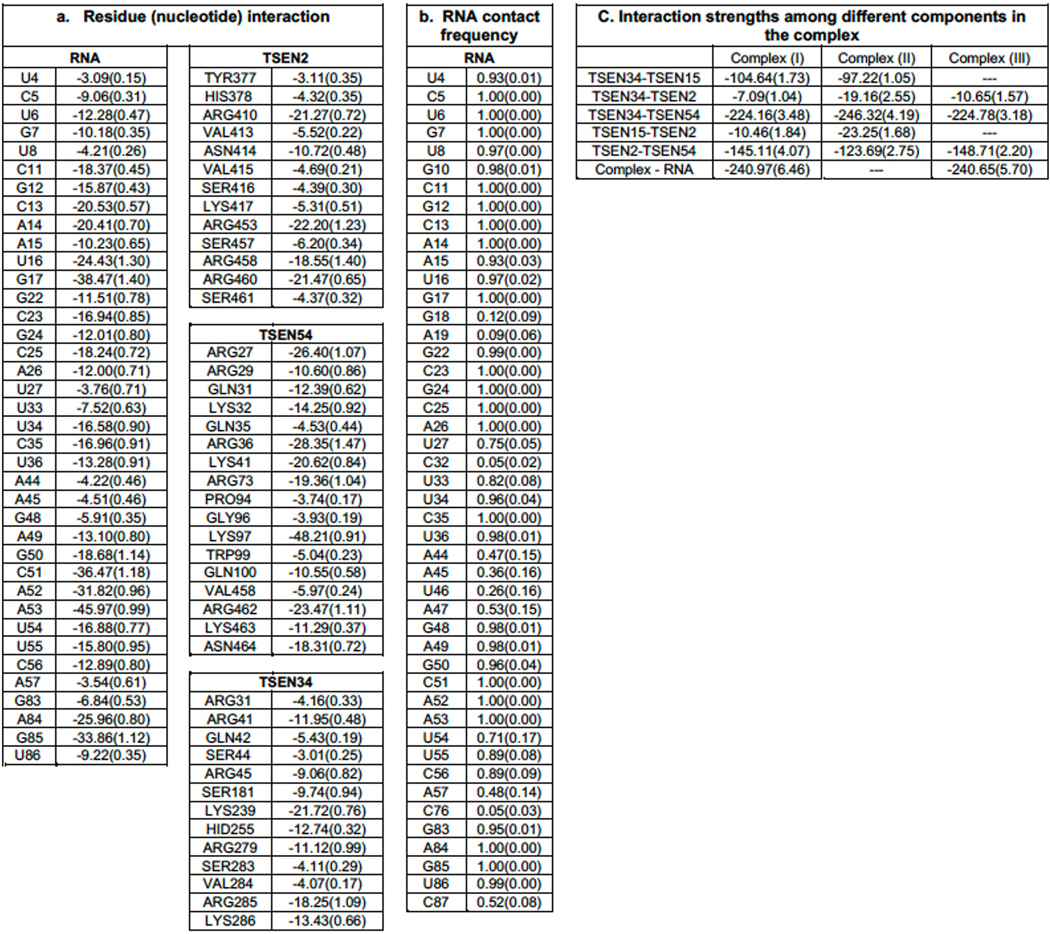
Extended Data Table. 1. Energy contributions and contact frequencies from molecular dynamic simulations.
a. MMGBSA energy calculated from 5000 configurations collected from 5 MD runs. The standard errors are given in parenthesis. Only energies with magnitudes greater than 3 kcals/mol are shown. b. Contact frequencies of nucleotides with the TSEN complex. The frequencies were calculated from 5000 configurations and if any heavy atom of a nucleotide is within 3.2 A to any heavy atoms of the protein complex, then the nucleotide is selected to be in contact with the complex. Only 35 out of 80 nucleotides are in contact 75% or more of the time of the simulation. c. interaction energies among various components in each system, calculated using MMGBSA.
|
