Figure 1: Structure and assembly of CRL3s.
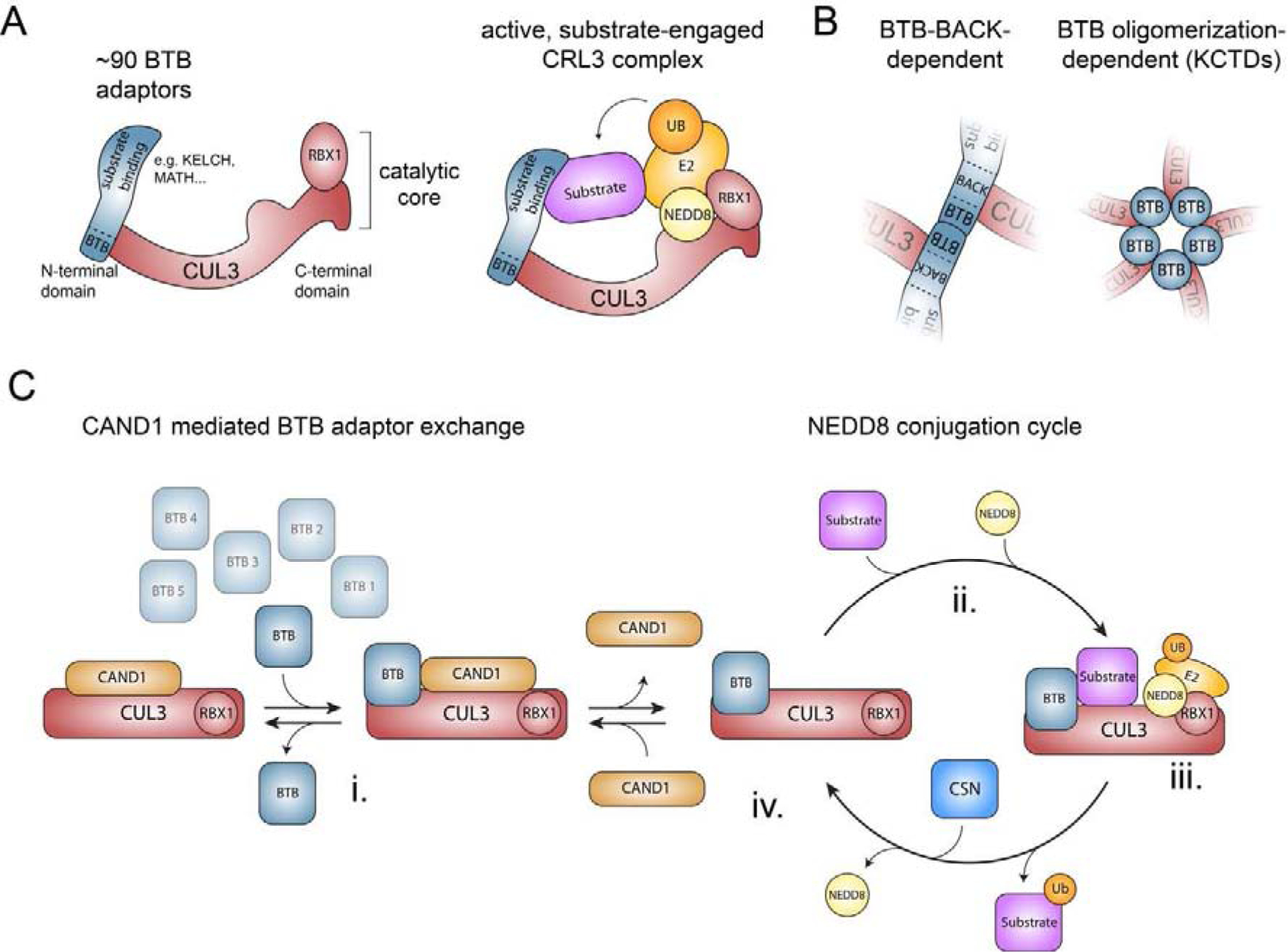
A) Schematic of the structure of a multi-subunit CRL3 complex. Left panel depicts a CRL3 that consists of CUL3 and the RING domain containing protein RBX1 (catalytic core) and one of ~90 interchangeable BTB proteins, which use their BTB domain to bind to the N-terminal domain of CUL3 and variable protein interaction domains to recruit specific sets of substrates. Right panel illustrates a model of an active, substrate-engaged CRL3 complex. Modification of CUL3 with NEDD8 at Lys-712 induces conformational changes and formation of ubiquitylation assemblies, in which RBX1 recruits an ubiquitin-charged E2 enzyme and the BTB protein position substrates for ubiquitin transfer. B) Schematic model of the two different modes how BTB proteins can interact with CUL3. C) Model of how CAND1-mediated BTB adaptor exchange and reversible NEDD8 modification are thought to regulate the cellular CRL3 repertoire (see text for details).
