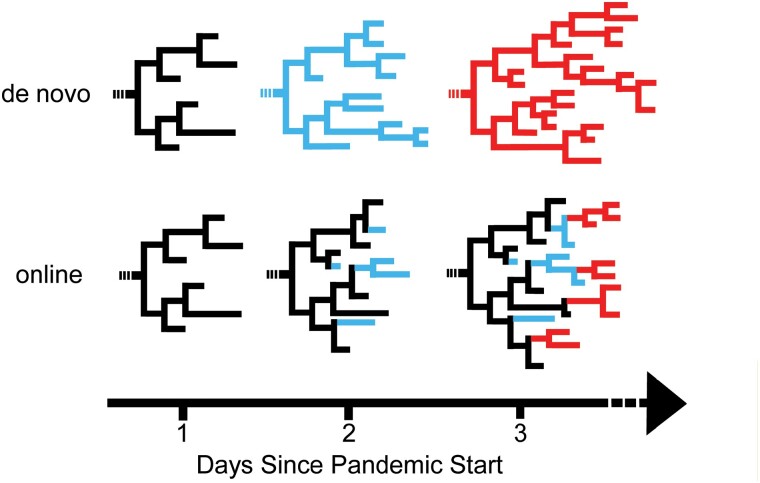
Figure 1.
Phylogenies may be optimized from scratch using de novo phylogenetics or iteratively using online phylogenetics. In de novo phylogenetics (top), trees are repeatedly re-inferred from scratch. Conversely, online phylogenetics (bottom) involves placement of new samples as they are collected. Online methods may be particularly well suited to pathogen datasets like SARS-CoV-2 where close relatives of ancestral samples already exist in the phylogeny. Intermittent optimization steps (not depicted) after new samples are placed can help overcome errors from previous iterations. Online phylogenetics is expected to be much faster and require less memory than de novo phylogenetics.