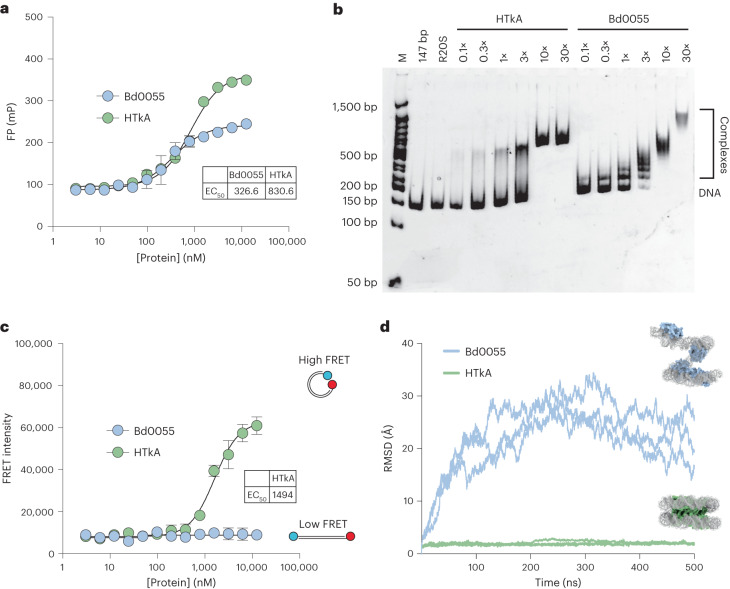
Fig. 4. Bd0055 binds DNA differently from HTkA.
a, Binding of Bd0055 to an Alexa488-labelled piece of 147 bp DNA (Widom 601 sequence), measured by FP (N = 3). EC50, half-maximal effective concentration. Mean ± s.d. b, Bd0055–DNA binding monitored by EMSA (using unlabelled 147 bp DNA). Bd0055 binds with similar affinity as HTkA, but produces a different gel shift pattern. c, Bd0055 was titrated into dual-labelled 147 bp DNA (Alexa488 and Alexa647, same DNA as in FP experiment). At concentrations of HTkA high enough to bind DNA, histones wrap the DNA and produce a high FRET signal. Bd0055 does not cause FRET between DNA ends, even with a large excess of protein (N = 3). Mean ± s.d. d, Plot of RMSDs from simulating a hypothetical Bd0055 hypernucleosome modelled onto PDB 5T5K using four histone dimers and 118 bp of DNA (see Supplementary Movie 1). After 500 ns of simulation, hypernucleosomes with HMfB remain stable, whereas hypothetical Bd0055 hypernucleosomes fall apart after as little as 10 ns.