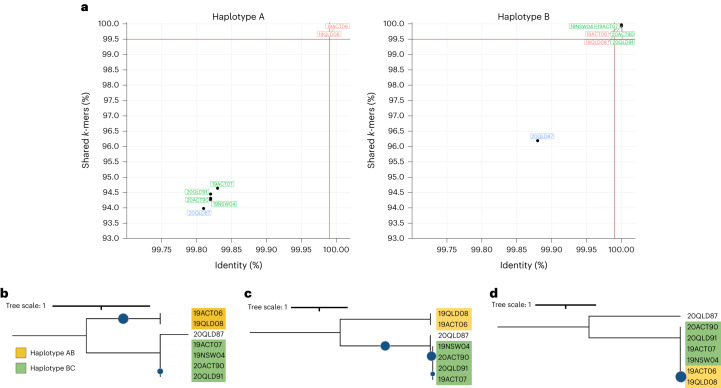
Fig. 1. Three distinct lineages and four haplotypes are present in a collection of seven Australian Pt isolates.
a, k-mer genome containment scores of Illumina sequencing reads against the Pt76 (19ACT06) haplotypes. Identity is the percentage of bases that are shared between the genome and the sequencing reads. Shared k-mers is the percentage of k-mers shared between the genome and the sequencing reads. Two red lines indicate thresholds above which we consider a haplotype genome to be fully contained in the sequencing reads of an isolate (identity ≥99.99%, shared k-mers ≥99.5%). The A haplotype is fully contained in the sequencing reads of two isolates (19ACT06, 19QLD08) and the B haplotype is fully contained in the sequencing reads of 6 isolates (19ACT06, 19QLD08, 19ACT07, 19NSW04, 20QLD91, 20ACT90). The 20QLD87 isolate contains neither A nor B haplotypes. b, The phylogenetic tree against the combined haplotypes 19ACT06 A and B indicate three lineages. c, The phylogenetic tree against the 19ACT06 haplotype A shows that two isolates share the A haplotype. d, The phylogenetic tree against the 19ACT06 haplotype B shows that six isolates share the B haplotype. Bootstrap values of over 80% are indicated with blue circles.