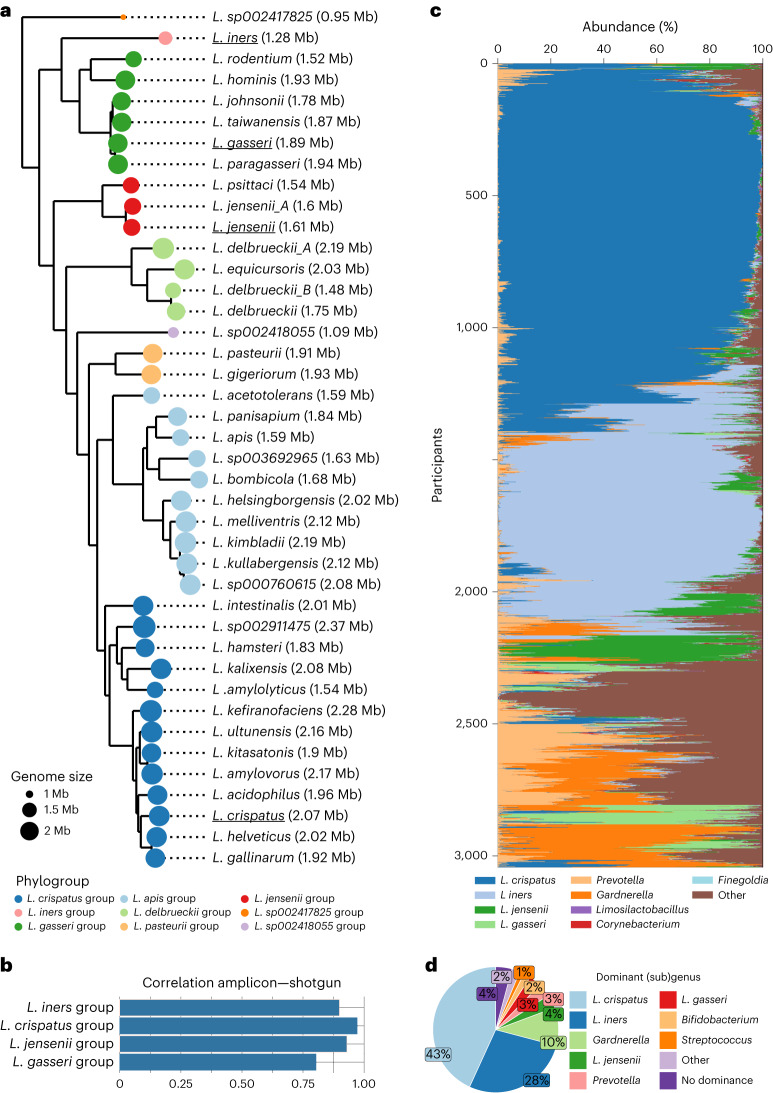
Fig. 2. Overview of the most abundant taxa in the vaginal microbiome of the Isala cohort with focus on the Lactobacillus taxa.
a, Maximum-likelihood phylogeny of species of the genus Lactobacillus inferred from the amino acid sequences of 100 single-copy core genes. Colours indicate the nine defined subgenera used in this study. Bold tip labels indicate representative species of the subgenera. Species names were taken from the Genome Taxonomy Database48, which splits species that are very diverse, yielding, for example, L. delbrueckii_A and L. jensenii_A, the latter recently identified as L. mulieris69. The size of the circles reflects the genome size (with the average genome size also shown between brackets). The underlined species names are the species names of the four typical vaginal Lactobacillus species, after which the subgenera are named. b, Validation of the 16S amplicon sequencing pipeline, including classification to Lactobacillus subgenera, with shotgun sequencing data (n = 18, Supplementary Fig. 2). For the typical four Lactobacillus subgenera, the Spearman correlations between their relative abundances in the amplicon and shotgun samples are shown. c, Stacked bar chart describing the microbiome composition of the Isala participants in terms of the nine taxa being first or second most abundant in at least 100 samples. d, Occurrence of the most dominant taxa based on the highest taxonomic resolution possible. Dominance was defined as the most abundant taxon that constituted at least 30% of the profile. ‘Other’ refers to the number of samples with a different (sub)genus dominant; ‘No dominance’ refers to the number of samples where no single (sub)genus reached at least 30% relative abundance.