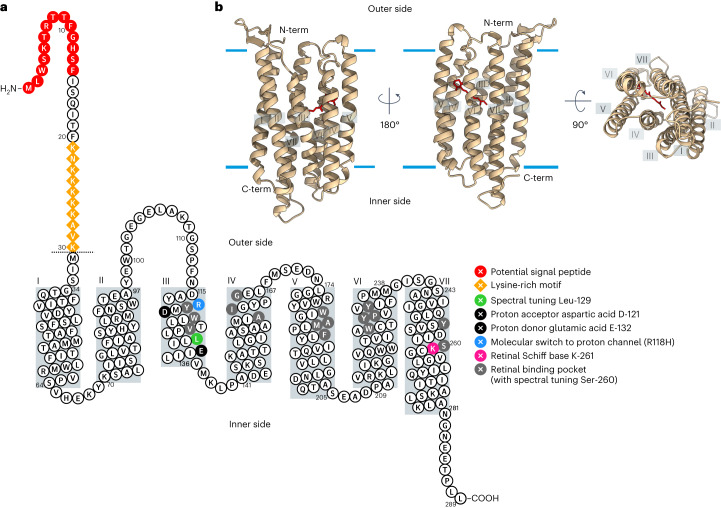
Fig. 1. Identification and in silico analysis of F. cylindrus rhodopsin 1 (FcR1).
a, Secondary structure prediction of FcR1 with key residues highlighted: proton acceptor D-121 and donor E-132 (black), K-261 (pink) forming a Schiff base link with retinal, and retinal binding pocket (Y-119, W-122, V-126, L-129, A-160, I-161, G-165, W-181, A-184, M-185, F-188, W-226, Y-229, P-230, Y-233, Y-253 and S-260) (light grey). The N-terminal extension of 30 amino acids of FcR1 compared to FcR2 is indicated as a dotted line. Potential N-terminal chloroplast signal peptide with conserved ‘ASAFAP’ (red circles) and lysine-rich motif (orange diamonds) in the N terminus of FcR1 are highlighted. Grey boxes correspond to transmembrane helices as predicted by AlphaFold2 structure prediction in b. b, Overall 3D view of the FcR1 protomer in the membrane as predicted by AlphaFold2. The experimental structure of PDB entry 4HYJ42 was used for structural alignment with FcR1 to position the retinal ligand, and the final model was generated by removing the structure of PDB entry 4HYJ. Estimated hydrophobic–hydrophilic membrane boundaries are shown as light blue lines.