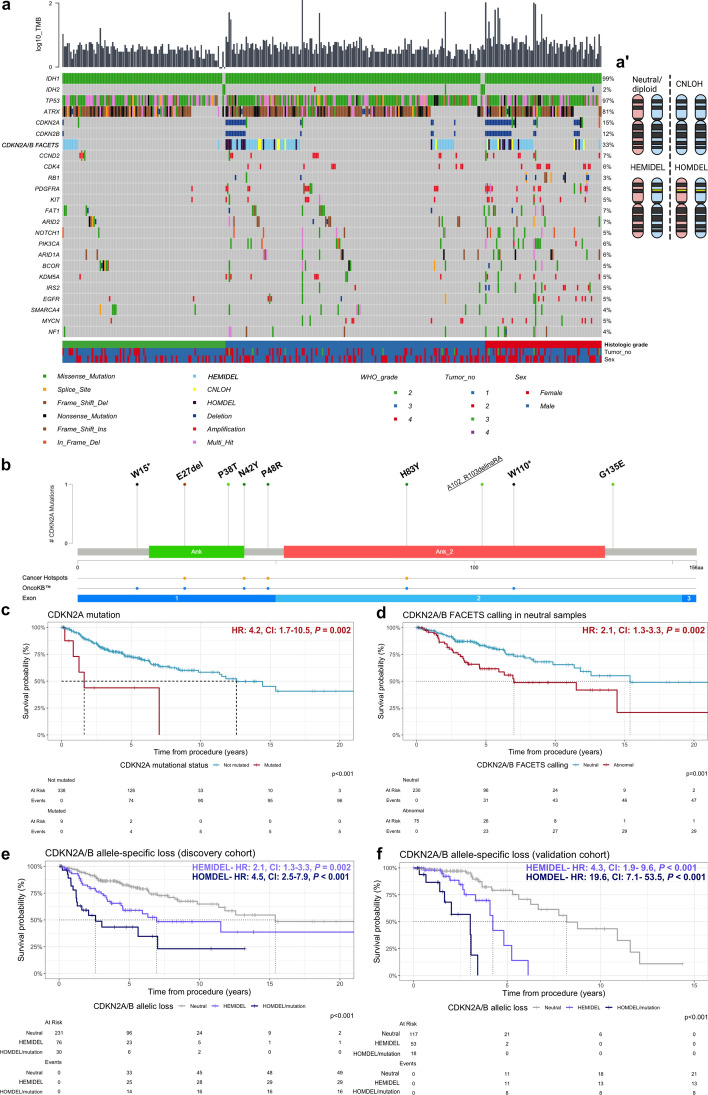
Fig. 1.
a Oncoplot depicting frequently altered genes and log10 tumor mutational burden (TMB) in 347 IDHA. a' CDKN2A/B allele-specific copy number states at 9p21.3. Yellow band = CDKN2A/B deletion, red and blue shades discern parental alleles. b Lollipop plots display variants in copy neutral CDKN2A/B (underlined: hypermutant). c Nonsynonymous CDKN2A mutations shorten OS. d ASCNA in CDKN2A/B copy neutral samples conferred worse OS than FACETS neutral. e CDKN2A/B HEMIDEL (n = 76) confers intermediate OS between patients with CDKN2A/B HOMDEL/mutation and CDKN2A/B neutral/without mutation. f TCGA validation cohort; HR—hazard ratios, CI—confidence intervals, P values within graphs refer to univariable Cox-proportional hazards regression models, log-rank test