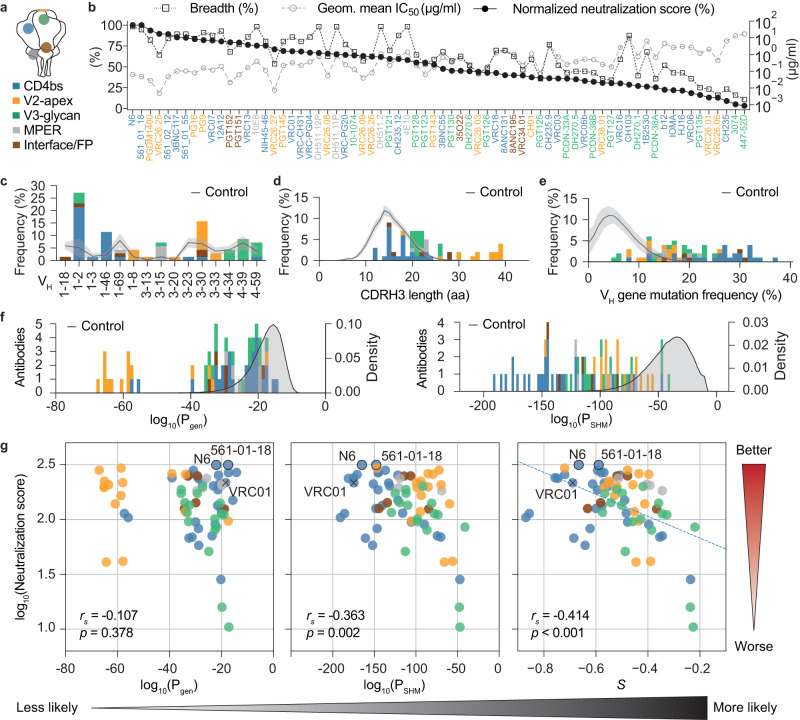
Fig. 2. Neutralization efficacy and sequence characteristics of broadly neutralizing antibodies targeting HIV-1.
a bNAb epitopes on the HIV-1 envelope spike. MPER membrane proximal external region, CD4bs CD4 binding site, FP fusion peptide. b Breadth, geometric mean half inhibitory concentration (IC50) of neutralized strains, and normalized neutralization score for n = 70 bNAbs that have been tested against the same 56 HIV-1 strains. c Heavy chain V gene segment usage for the selected bNAbs. d Heavy chain CDR3 length distribution for the selected bNAbs in amino acids (aa). e Heavy chain V gene mutation frequency distribution for selected bNAbs. Controls in c–e depict the mean frequencies of the respective sequence features as solid lines ± SD as shaded areas from the n = 57 uninfected individuals (Fig. 1 f–h). f Heavy chain Pgen and PSHM distribution for selected bNAbs derived through IGoR by a model learned on productive sequences from n = 57 uninfected individuals. Controls represent Pgen and PSHM distributions for productive sequences from the n = 57 uninfected individuals. g Correlation plots of bNAb neutralization scores against heavy chain Pgen, PSHM, and a combined probability score S = c1log10(Pgen) + c2log10(PSHM), which was derived by a linear regression (dashed line) with c1 = 3.248 × 10−03 and c2 = 3.604 × 10−03. Spearman correlation coefficients r and two-sided p values are given in the figure. The correlation coefficient and two-sided p value from the linear regression for S are r = −0.468 and p = 4.392 × 10−05. Two near pan-neutralizing antibodies (N6 and 561-01-18) are highlighted by black outlines, one antibody that has been used for structure-guided vaccine approaches (VRC01) is highlighted by ‘x‘. Gradients on the right and bottom show directions for increasing neutralization activity and generation probability, respectively. Source data are provided as a Source Data file.