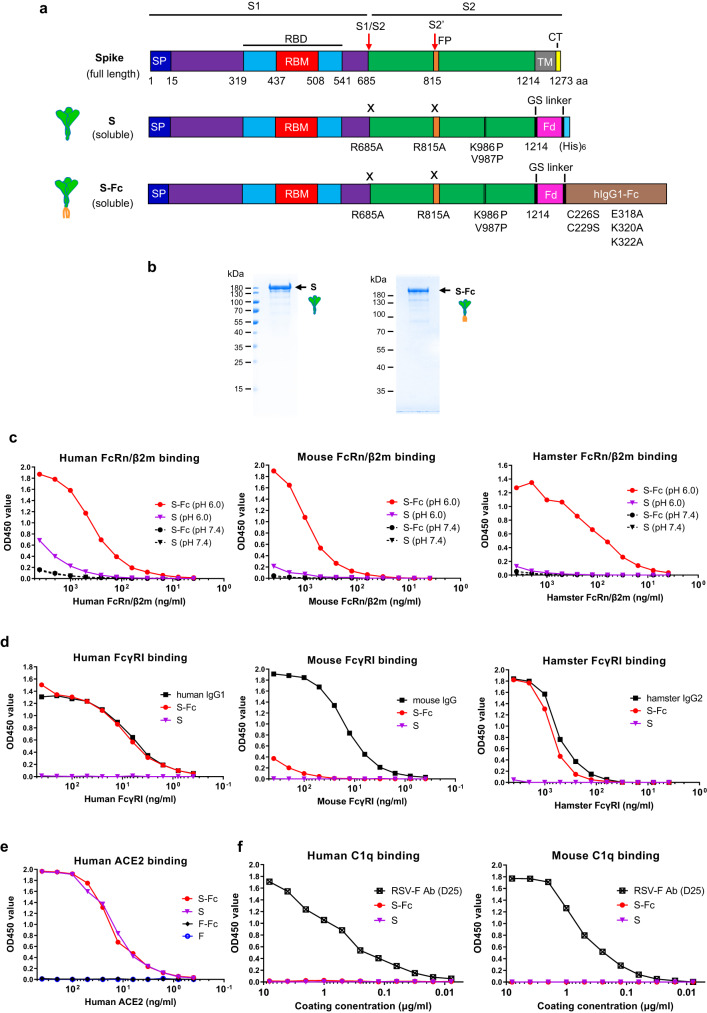
Fig. 1. Expression and characterization of the Spike and prefusion Spike-Fc (S-Fc) protein.
a Schematic illustration of the full-length protein sequence of SARS-CoV-2 Spike. SP: signal peptide; RBD: receptor binding domain; TM: transmembrane domain; CT: cytoplasmic tail. Graphic design of the fusion of SARS-CoV-2 Spike with the T4 fibritin foldon domain (Fd) to create a soluble, prefusion-stabilized, and trimeric Spike protein. Mutations were made in the SARS-CoV-2 (strain USA/WA1/2020) spike by replacing Arg 685 and Arg 815, respectively, with an Ala residue to remove the cleavage site and replacing Lys 986 and Val 987, respectively, with a Pro residue to create a prefusion-stabilized form. The diagram demonstrates the fusion of a SARS-CoV-2 Spike with the T4 fibritin foldon domain and human Fcγ1 to create a prefusion-stabilized and trimeric S-Fc fusion protein. Mutations were also made in the Fcγ1 fragment by replacing Cys 226 and Cys 229, respectively, with a Ser residue to abolish Fc dimerization, and replacing Glu 318 Lys 320 Lys 322 with Ala residues to remove the complement C1q binding site. b The S and S-Fc fusion proteins were purified from the stable CHO cell lines. The soluble S and S-Fc proteins were purified by anti-His and Protein A affinity chromatography, respectively, subjected to SDS-PAGE gel electrophoresis under reducing conditions and visualized with Coomassie blue staining. The figure is a representative result from three independently-repeated experiments. c–f Test of the S-Fc binding to human, mouse, or hamster FcRn/β2m; human, mouse, or hamster FcγRI; human ACE2; and human or mouse C1q. The ELISA determined the specific binding. The purified S protein was used as a positive control for ACE2 binding and a negative control for FcRn/β2m or FcγRI binding. Respiratory syncytial virus (RSV) protein F alone or Fc-fused F proteins were used as a negative control. Mouse IgG, human IgG1, hamster IgG2, and a mAb (D25) against RSV F protein were used as the positive control, respectively. Source data are provided as a Source data file.