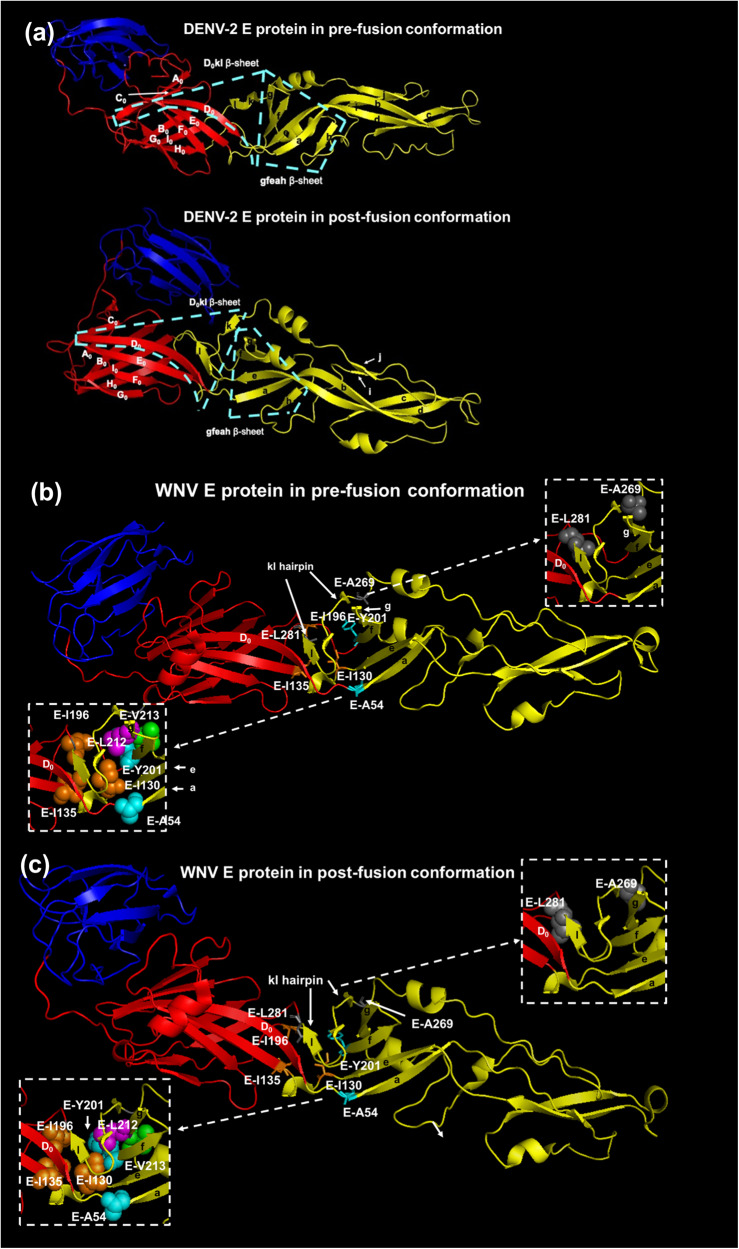
Fig. 5. Six flavivirus-conserved hydrophobic residues crucial for the structure–function of the E protein.
The two β-sheets at the EDI-EDII interface are highlighted in the pre-fusion and post-fusion conformations of DENV-2 E protein with cyan dash line in (a) because the crystal structure of WNV E protein is only available in pre-fusion conformation. The three domains of the E protein are colored in red (EDI), yellow (EDII), and blue (EDIII). The six flavivirus-conserved residues in the EDI-EDII hinge region of the pre-fusion (b) and post-fusion (c) conformations of WNV E protein are shown with stick (PDB ID:2HG0). The post-fusion conformation of WNV E protein is predicted using the post-fusion conformation of St. Louis encephalitis virus E protein (PDB ID: 4FG0). The detailed view highlighted the location of the gfeah and D0kl β-sheets. The E-A54 and E-Y201 residue at the N-terminus of the a and f β-sheets are highlighted with cyan spheres, respectively. The E-Y201 residue is proximal to the flavivirus-conserved E-L212 (green) and E-V213 (magenta) residues in the fg hairpin motif. The E-I130, E-135, and E-I196 residues are labeled with orange spheres. The E-A269S and the E-L281N mutation in the fourth motif of the EDI-EDII hinge region are shown in gray and are part of the klD0 β-sheet.