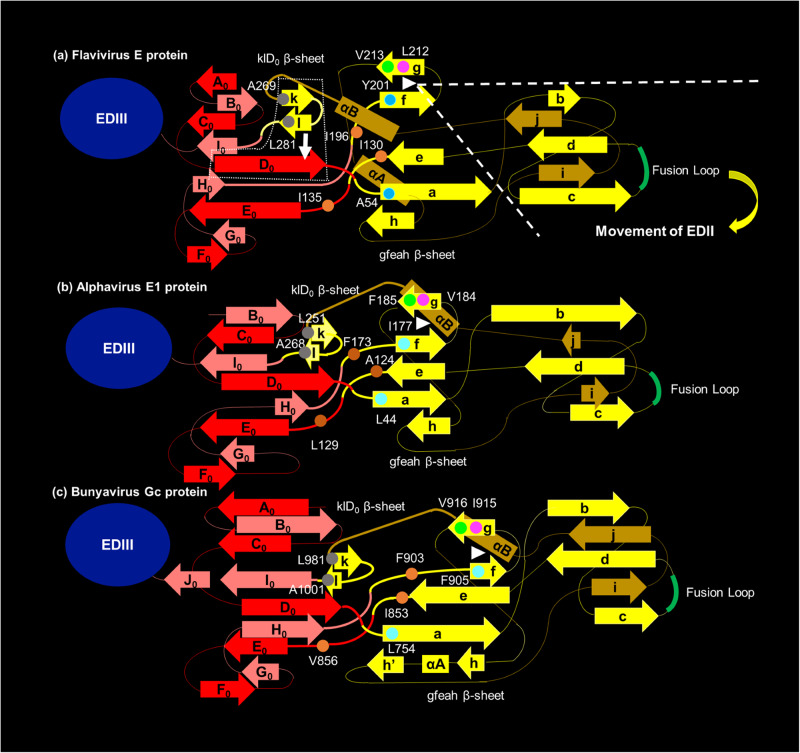
Fig. 6. Proposed model for the arbovirus conserved hinge effect to control the EDI-EDII interdomain movement of E protein.
a represents the proposed role of each flavivirus-conserved hydrophobic residue in exerting the hinge effect at the EDI-EDII interface of E protein. For simplicity, EDIII independent from the EDI-EDII interdomain movement is shown in a blue oval. The four motifs of the EDI-EDII hinge region are highlighted in thick lines. The WNV E-A54 and E-Y201 residues are at the base of the gfeah β-sheet, reflecting their importance in the relative movement of EDII in relation to EDI. The E-Y201 residue and the flavivirus-conserved E-L212 (magenta) and E-V213 (green) residues form the fg hairpin (white triangle). The gfeah β-sheet rotates along the fg hairpin to change the orientation of EDII (highlighted by the white dash line). The movement of EDII triggers the rotation of A0C0D0E0F0 β-sheet of the EDI. The new conformation of the D0 β-strand is stabilized by the hydrogen bonds with the l β-strand separated from the kl hairpin structure of EDII (indicated by the white arrow). The eqivalent hydrophobic residues in the alphavirus E1 proteins and bunyavirus Gc proteins are predicted using the structural alignment function of Modeller and Pymol software packages, reflecting that there is a putative common mechanism underlying the conformational change of the class II fusion proteins from dimer to trimer. The numbering is based on Semliki Forest virus (SFV) E1 protein (b) and Rift Valley fever virus (RVFV) Gc protein (c). The fg hairpin of alphavirus E1 protein and bunyavirus Gc protein (white triangle) both consist of three conserved hydrophobic amino acids. Specifically, the base of the fg hairpin motif at the EDI-EDII hinge region of SFV involves the alphavirus-conserved SFV E1-I177 (cyan), E1-V184 (margenta), and E1-F185 (green) hydrophobic residues. The fg hairpin motif is proximal to three alphavirus-conserved SFV E1-A124, E1-L129, and E1-F173 hydrophobic residues (shown in orange) equivalent to WNV E-I130, E-I135, and E-I196 residues, respectively. The base of the a β-strand of SFV E1 protein consists of the alphavirus-conserved E1-L44 residue (cyan), equivalent to the WNV E-A54 residue. And, the kl hairpin of the SFV E1 protein is potentially stabilized by two alphavirus-conserved hydrophobic amino acids, including the E1-L251 and E1-A268 residues (shown in gray), resembling the WNV E-A269 and E-L281 residues, respectively. Similarly, the fg hairpin motif at the EDI-EDII hinge region of RVFV Gc protein may constitute the bunyavirus-conserved RVFV Gc-F905 (cyan), Gc-I915 (margenta), and Gc-V916 (green) residues. The core of the RVFV Gc protein EDI-EDII hinge region also contains three bunyavirus-conserved hydrophobic amino acids, including the RVFV Gc-W855, Gc-L859, and Gc-F903 residues (show in orange), which resemble the relative location of the WNV E-I130, E-I135, and E-I196 residues, respectively. The close proximity between flavivirus-conserved WNV E-A54 and I130 residues is followed by the analogous RVFV Gc-L754 (cyan) and Gc-W855 (orange) residues, respectively. The kl hairpin of the RVFV Gc protein is potentially stabilized by the Gc-L981 and Gc-A1001 residues (shown in gray) conserved across different bunyaviruses.