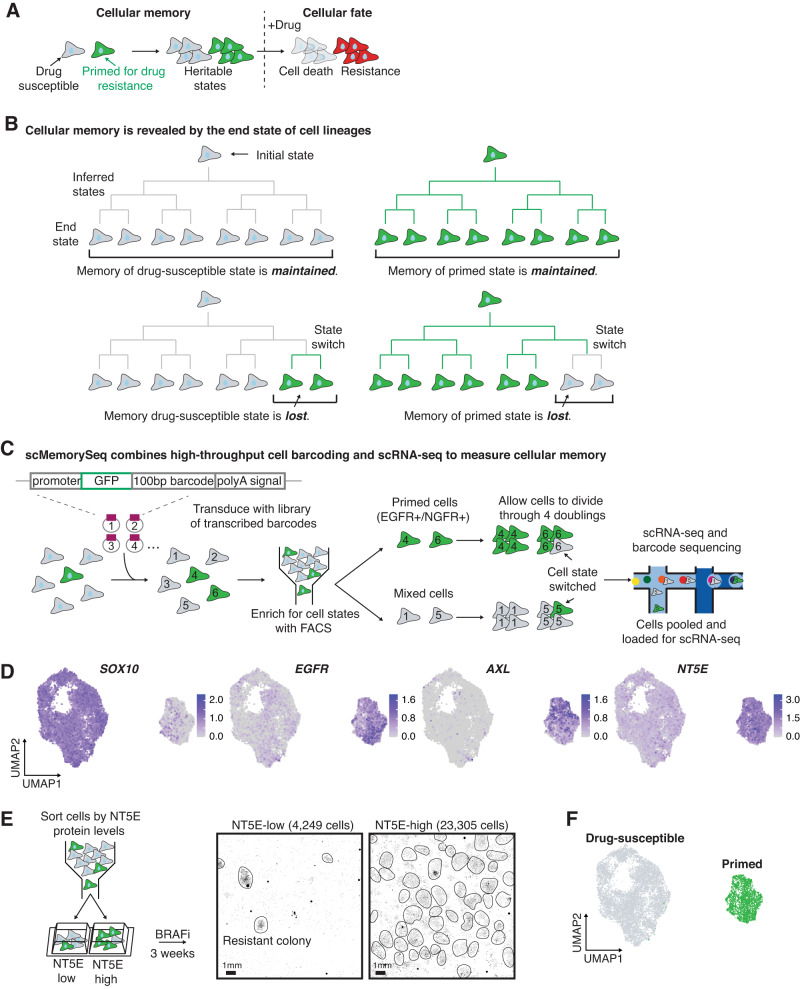
Fig. 1. scMemoryseq can reveal when cells change state.
A Model for how the heritable primed state leads to resistance to targeted therapy. Cells primed for drug resistance (in green) proliferate and pass on their gene expression state through cell division, which demonstrates the concept of gene expression memory. These primed cells survive treatment with BRAFi and MEKi (resistant cells in red) while the drug-susceptible cells (in gray) die. B Schematic of lineages that maintain memory compared to those that lose memory. In lineages that maintain memory, the end state of all the cells is the same as the initial state. In lineages where memory is lost, the end state contains a mixture of cell states. C Schematic of the experimental design we used to capture the transcriptome and lineage of cells. We transduced melanoma cells (WM989) with a high-complexity library of lentiviral lineage barcodes. We then sorted a sample of primed cells (based on EGFR and NGFR) and a mixed population. This sorting step provides us with the initial state of the cells. We then allowed the cells to undergo approximately four doublings before scRNA-seq and barcode sequencing on the cells. D UMAP plots showing the log10 normalized gene expression of the drug-susceptible cell marker SOX10, and the primed cell markers EGFR, AXL, and NT5E. E Diagram of the experimental design and images showing the fixed NT5E-low and NT5E-high cells stained with DAPI after 3 weeks in targeted therapy. Each black dot is the nucleus of a drug-resistant cell, and drug-resistant colonies are circled in black, n = 3 biological replicates. F UMAP plot showing drug-susceptible cells (in gray) and primed cells (in green).