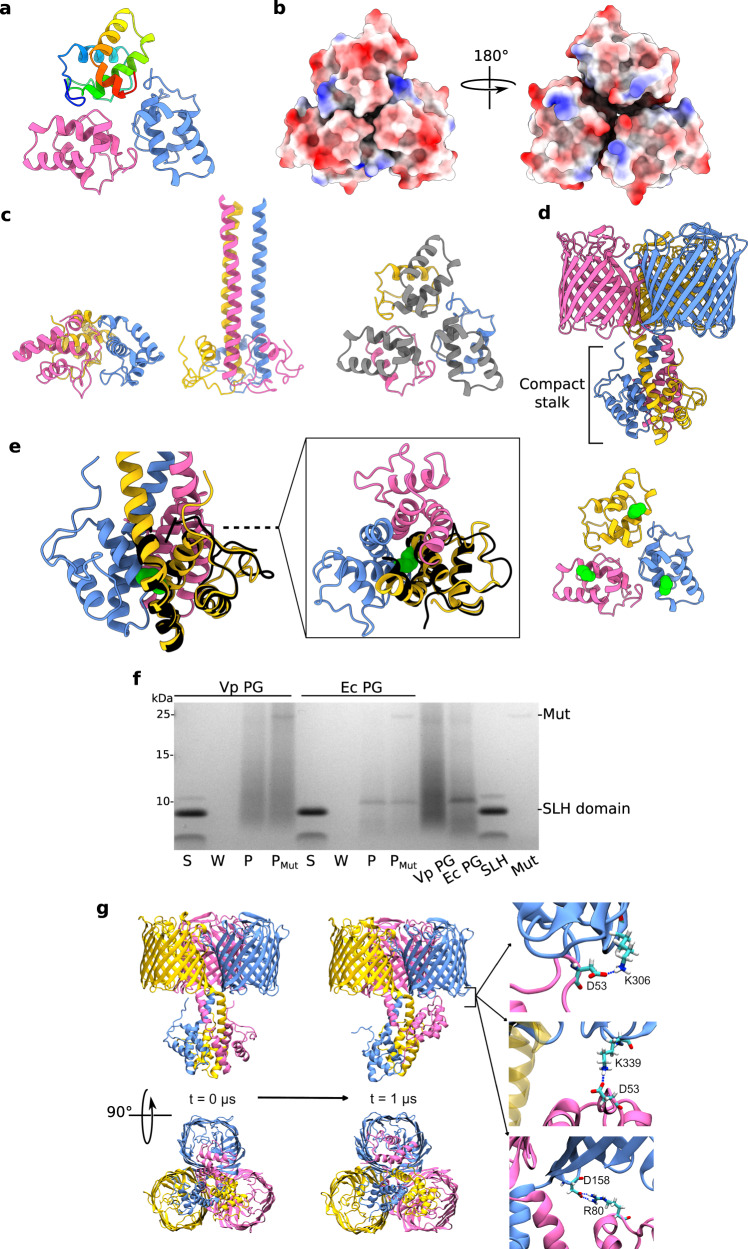
Fig. 4. Crystallography, AlphaFold2, and binding assay show evidence of alternate stalk conformations.
a Crystal structure of VpOmpM1 stalk trimer at 1.7 Å. Residues 23–105 were resolved. One chain is in rainbow: N-terminus is blue, C-terminus is red. b Electrostatic surface presentation of the stalk crystal structure. (Left) Putative view from the OM and (right) from the periplasm. c Comparison of the stalk crystal structure (left) and AlphaFold2 predicted extended conformation (middle). Only residues 23–105 are displayed for both. (Right) Residues 64–105 of the stalk crystal structure that form the extended coiled-coil in the AlphaFold2 prediction are in grey. d Alternative, compact stalk conformation predicted by AlphaFold2. e Close-up view of the compact stalk prediction with a single chain from the crystal structure (black) superposed (Cα-Cα r.m.s.d. 0.78 Å) (left). (Middle) Top view down the section marked by the dashed line. (Right) Stalk crystal structure. The sidechain of M75 is shown in green space-filling representation in each panel. f Binding assay of recombinant VpOmpM1 SLH domain to sacculi (PG) isolated from V. parvula and E. coli. After incubation of protein with PG, the insoluble PG was pelleted by centrifugation, washed by resuspending in buffer, and pelleted again. The PG was then resuspended and split into two aliquots, one of which was incubated with mutanolysin (Mut). The supernatant from the binding reaction (S), the wash (W) and the resuspended PG pellet without (P) and with Mut treatment (PMut) were boiled and analysed by SDS-PAGE. Three independent binding experiments were performed and yielded similar results. g All-atom MD simulation of AlphaFold2 predicted compact stalk model over 1 μs. The SLH domain of one of the protomers (pink) reaches towards the OM and interacts with the periplasmic turns of the β-barrel of another protomer (blue). The salt bridges shown on the right had occupancies of 5.8%, 3.44% and 1.89% (top to bottom) throughout the simulation.