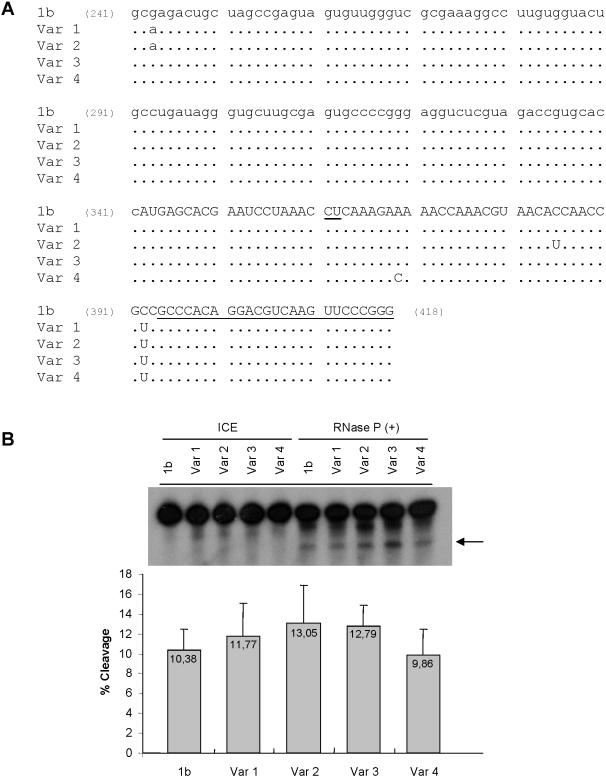
Figure 3.
RNase P sensitivity of HCV RNA natural variants bearing substitutions in the RNase P cleavage region. (A) Sequence alignment of nucleotides 241–418 of cloned HCV variants. 1b represents the genotype 1b reference sequence and variants 1–4, four HCV variants isolated through cloning and sequencing of 2–418 region of HCV RNA (lower case, HCV 5′ NCR, upper case, HCV core coding region). RNase P cleavage site is underlined at positions 361–363. The position of the 3′ primer used for PCR and cloning is underlined (394–418). (B) Autoradiogram of RNase P in vitro cleavage assay of labeled 2–418 HCV RNAs. Variants 1–4 were incubated with RNase P, in parallel to 1b HCV RNA control, 1 h 30 min at 30°C. ICE indicates the input RNA, not incubated. Reaction products were fractionated in denaturing polyacrylamide–UREA gel electrophoresis followed of X-ray film exposure. The arrow indicates the larger cleavage product. Experiment was repeated as a triplicate and quantified with a Radioisotopic Image Analyser FLA-5000 (Fuji). The table below the autoradiogram indicates the average cleavage of each RNA determined as the ratio: products/products + uncleaved RNA.