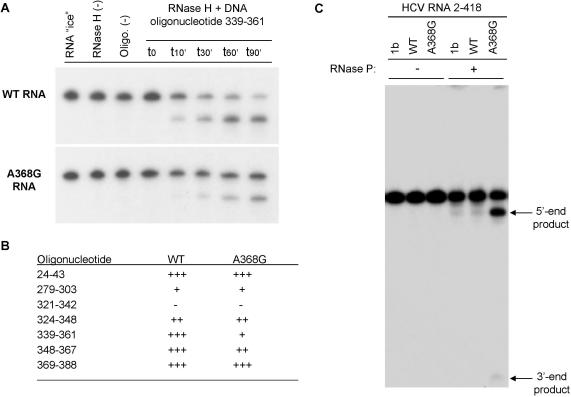
Figure 4.
Enhanced accessibility to RNase P of the in vitro selected HCV RNA variant partially resistant to RNase H. (A) Time course DNA-mediated RNase H cleavage of the [α-32P]GTP-labeled WT RNA compared to the A368G variant RNA (nucleotides 2–418), selected as indicated in Figure 2 using a DNA oligonucleotide complementary to nucleotides 339–361 of HCV RNA. Products were analyzed by denaturing PAGE (4%) and visualized by autoradiography. The kinetic study (0–90 min) shows the resistant phenotype associated with the mutant natural variant (estimated at a 3-fold effect). (B) Relative sensitivities of WT and A368G 2–418 HCV RNA to DNA-mediated RNase H cleavage. The oligonucleotide column indicates the positions where DNA oligonucleotides hybridize to RNA. +++: 75–100% of cleavage after 90 min, ++: 50–75%. +: 25–50%, −: 0–25%. (C) In vitro human RNase P cleavage of HCV 2–418 RNA. Lane 1b is the control genotype 1b HCV clone previously used in the laboratory to study RNase P cleavage (10) and represents here the cleavage efficiency and the product size control, lane WT is the HCV predominant sequence of the patient studied here, and lane A368G is the selected mutant sequence. RNase P (+) and (−) are, respectively, reactions with and without RNase P. Products were analyzed by denaturing PAGE (4%) and visualized by autoradiography. The position of the two 5′ and 3′ end RNase P cleavage products is indicated on the right.