Abstract
Follicular lymphoma is one of the most frequent lymphomas. Histologically, it is characterized by a follicular (nodular) growth pattern of centrocytes and centroblasts; mixed with variable immune microenvironment cells. Clinically, it is characterized by diffuse lymphadenopathy, bone marrow involvement, and splenomegaly. It is biologically and clinically heterogeneous. In most patients it is indolent, but others have a more aggressive evolution with relapses; and transformation to diffuse large B-cell lymphoma. Tumorigenesis includes an asymptomatic preclinical phase in which premalignant B-lymphocytes with the t(14;18) chromosomal translocation acquire additional genetic alterations in the germinal centers, and clonal evolution occurs, although not all the cells progress to the tumor stage. This manuscript reviews the pathobiology and clinicopathological characteristics of follicular lymphoma. It includes a description of the physiology of the germinal center, the genetic alterations of BCL2 and BCL6, the mutational profile, the immune checkpoint, precision medicine, and highlights in the lymphoma classification. In addition, a comment and review on artificial intelligence and machine (deep) learning are made.
Keywords: follicular lymphoma, pathobiology, pathogenesis, prognosis, mutational profile
Subtypes of follicular lymphoma
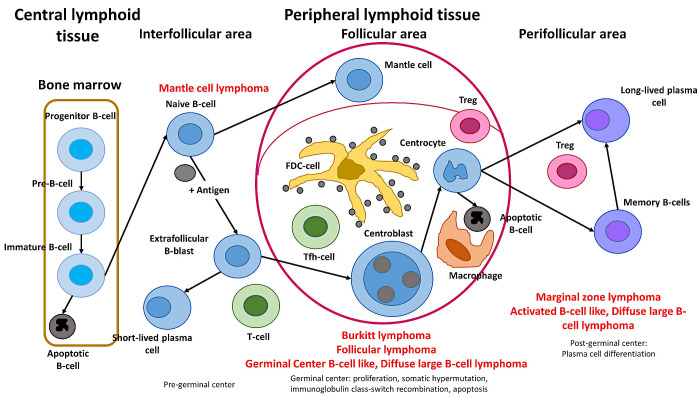
The lymphoma classification of lymphoid neoplasms, as revised in 2017, included more than 80 mature lymphoid neoplasms that were divided into three main subtypes: Hodgkin, B-cell, and T-cell lymphomas.1,2 The broad categories of mature B-cell neoplasms are shown in Figure 1. In this classification, each lymphoma subtype arises or has a stage of differentiation from a particular compartment of the immune system. Therefore, the classification is based in part on the physiological cell counterpart or cell of origin. For example, follicular lymphoma, diffuse large B-cell lymphoma, and Burkitt lymphoma would arise from the germinal centers.
Fig. 1.
Cell-of-origin and classification of mature B-cell neoplasms
The different lymphoma subtypes are classified based on their histological architecture, cellular morphology, immunophenotype, genetic characteristics, and correspondence to the normal stages of B-cell development, such as naïve, germinal center, and post germinal center cells. Follicular lymphoma comprises germinal center B-cells similar to those found in secondary lymphoid follicles. It is composed of a mixture of small (centrocytes) and larger neoplastic cells (centroblasts) that have the t(14;18)/IGH::BCL2 fusion gene.
Follicular lymphoma is one of the most frequent lymphomas. Follicular lymphoma represents approximately 5% of all hematological neoplasms, and around 20 to 25% of all new non-Hodgkin lymphomas in western countries.3 As reported by Yoshino T. et al., the incidence of lymphoma has rapidly increased over the last 40 years in Japan, reaching a frequency comparable to that of Western countries.4
The follicular lymphoma category comprises the following entities: follicular lymphoma (including in situ follicular neoplasia, and duodenal-type follicular lymphoma), BCL2-R–negative CD23-positive follicle center lymphoma, primary cutaneous follicle center lymphoma, pediatric-type follicular lymphoma, and testicular follicular lymphoma.5-9
Structure and function of the germinal center
Figure 2 shows a simplified version of the dynamics of the germinal center. The germinal center of lymphoid follicles is the place where activated B-lymphocytes acquire the diversity of immunoglobulin genes using the somatic hypermutation (SHM) mechanism; as a result, high-affinity antibodies are created. In that specialized immune microenvironment, B-lymphocytes also undergo class switch recombination (CSR) to create antibodies with specialized effector functions.10
Fig. 2.
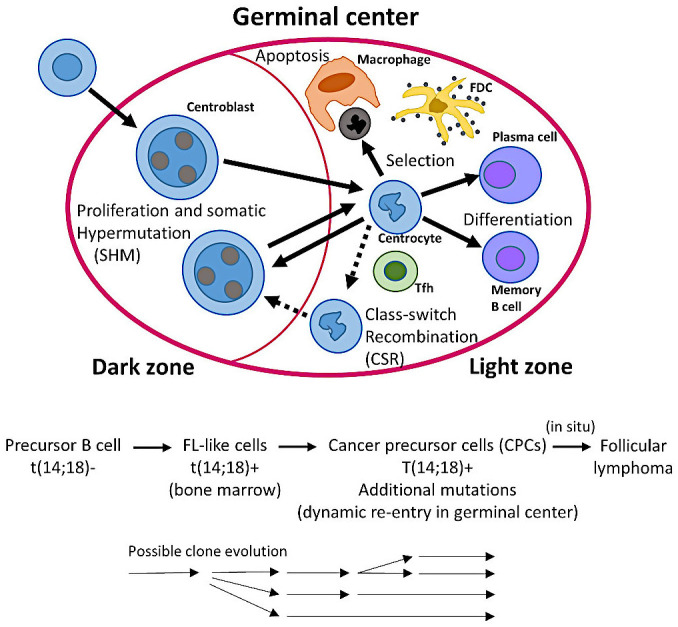
Dynamics of B-lymphocytes in the germinal centers; and a stepwise evolution model of follicular lymphoma
The germinal center reaction is crucial in humoral immunity, during which somatically mutated B-lymphocytes and plasma cells with high-affinity receptors are generated. The initiation and formation of mature germinal centers follows a series of phases including early initiation (antigen activation), late initiation (T cell migration into follicle, and appearance of early germinal centre), and proliferation and establishment (appearance of mature germinal center). Each phase is characterized by a different expression and/or functional requirement of molecules, NFKB and MYC are necessary in the initiation stages, while BCL6, SPIB, and BACH2 in late initiation and mature stages. This figure shows the dynamics of the germinal center reaction and selection of high-affinity antibody mutants. Antigen-activated B-lymphocytes undergo clonal expansion in the dark zone, during proliferation, somatic hypermutation (SHM) introduces basepair changes into the V(D)J region of the variable region (IgV) genes of immunoglobulin. In the light zone, with the help of follicular dendritic cells (FDCs) and T follicular helper (Tfh) cells, B-lymphocytes with improved-modified B-cell receptor (BCR) high-affinity are positively selected. Then, some will recirculate into the dark zone, undergo class switch recombination (CSR), or differentiate into plasma cells or memory B-lymphocytes. In the light zone, B-lymphocytes with unfavorable antibodies that are unable to capture enough antigen undergo apoptosis.
In the early immune response, in the border between the B-cell and T-cell zones, the antigen-activated B and T-lymphocytes are committed to differentiate into germinal center B-cells and T follicular helper (Tfh) cells. The migration into the follicle is facilitated by BCL6 in the case of B-cells, which happens after the Tfh cells have moved into the follicle. B-lymphocytes differentiate into blasts that proliferate until filling the follicle.10-14 When the germinal center matures, there is polarization into two different microenvironments, the dark and light zones. In the dark zone, B-lymphocytes proliferate extremely rapidly; and generate a large quantity of mutations in immunoglobulin (SHM). Then, dark zone B-lymphocytes differentiate into light zone B-lymphocytes. There, the ones with high-affinity antibodies are selected; and instructed to recirculate to the dark zone to undergo further SHM (process mediated by MYC, and REL); or to differentiate into memory or plasma cells.10-14 Follicular lymphoma is characterized by an immune microenvironment that includes all the components of the germinal center. The germinal center B-cell origin of follicular lymphoma is also supported by the identification of ongoing somatic hypermutation of the immunoglobulin heavy chain variable region (IGVH) of t(14;18)-positive lymphoma cells.15-19
Clinicopathological characteristics of follicular lymphoma
Follicular lymphoma originates or has a differentiation stage of germinal B-lymphocytes; and in most cases grows in a follicular (nodular) architectural pattern. Follicular lymphoma is clinically indolent in most cases and is associated with a favorable outcome. Nevertheless, in a fraction of patients, the disease behaves more aggressively with progression, and adverse outcomes. Because of the use of rituximab, outcomes of the follicular lymphoma patients have improved significantly, with around 80% of the patients having an overall survival above 10 years.3 Prognostic factors include the Follicular Lymphoma International Prognostic Index (FLIPI) that includes age, nodal sites, LDH, hemoglobin, and stage variables20 and is also valid in the rituximab era;21 the PRIMA prognostic index (PRIMA-PI) that uses B2-microglobulin,22,23 tumor grade (probably grade 1-3 A versus 3B),24-28 and microenvironment,29-49 among others.
From a clinical perspective, follicular lymphoma in adults and children is different. Pediatric-type follicular lymphoma is characterized by a low-stage disease (I/II), involvement of the head and neck region, high histological grade,3 absence of BCL2 rearrangement, mutations of MAP2K1 and TNFRSF14, and a high rate of cure.50-53
Pathogenesis
The pathogenesis of follicular lymphoma is multifactorial; and involves a series of steps in which a B lymphocyte acquires genetic and epigenetic alterations that lead to malignant transformation.54 The molecular changes can occur at different compartments of the immune system, including the bone marrow where BCL2 rearrangement occurs in precursor B-cells; or in the germinal centers of secondary lymphoid organs where somatic mutations and class switch recombination occurs.54
In the stepwise evolution model of follicular lymphoma,3 the evolution follows three steps: healthy or subclinical disease, primary tumor, and relapse (Figure 2). The t(14;18) occurs in the bone marrow. FL-like cells (FLLCs) are in most healthy individuals and are characterized by t(14;18). Follicular lymphoma emerges from early mutated cancer precursor cells (CPCs) engaging in a dynamic process of reentry into the germinal center, evolving and disseminating over decades in asymptomatic individuals.3 Such early clones are likely responsible for posttreatment relapses. In situ follicular neoplasia (ISFN) represents an early precursor lesion that may progress into follicular lymphoma at a low rate of progression.3
Follicular lymphoma neoplasia is characterized by a mixture of small cleaved B-lymphocytes (known as centrocytes); and larger noncleaved B-lymphocytes (known as centroblasts); and a tumor immune microenvironment that mimics the structure of the secondary follicles55 (Table 1). This dynamic structure comprises reticular cells, follicular dendritic cells (FDC), T follicular helper (Tfh) cells, FOXP3-positive T follicular regulatory (Tfr) cells, and macrophages.13 Table 1 shows the grading. The histological pattern is follicular (>75%), follicular and diffuse (25-75%), focally follicular/predominantly diffuse (<25%), and diffuse (0% of follicular).2
Table 1. Grading and mutational landscape of follicular lymphoma.
| Grading of follicular lymphoma | ||
|---|---|---|
| Grade | CB:HPF | Characteristics |
| 1 | 0-5 | CD10+, BCL2+, and BLC2 translocation+ in 90% of cases |
| 2 | 6-15 | CD10+, BCL2+, and BLC2 translocation+ in 90% of cases |
| 3A | >15 | Presence of centrocytes, CD10+, BCL2+, and BLC2 translocation+ in 75% of cases |
| 3B | >15 | Absence of centrocytes (diffuse areas of centroblasts), CD10+, BCL2+, and BLC2 translocation+ in few cases |
| Mutational profile | ||
| Gene | % | Function / Effect |
| KMT2D | 80-90 | Loss of function; histone modification |
| IgHV, IgLV | 75-90 | Gain of function; BCR signaling and proliferation |
| CREBBP | 33-70 | Loss of function; histone modification |
| BCL2 | 0.5 | Gain of function; anti-apoptosis |
| TNFRSF14 | 20-50 | Loss of function; immune evasion |
| BCL6 | 47 | Gain of function; tumor progression |
| H1-2, H1-4 | 44 | Loss of function; chromatin remodeling |
| RRAGC | 17 | gain of function; mTOR survival |
| EZH2 | 7-30 | Gain of function; histone modification |
| TNFAIP3 | 2-26 | Loss of function; survival |
| Prognosis | ||
| m7-FLIPI | EZH2, ARID1A, MEF2B, EP300, FOXO1, CREBBP and CARD11 | |
| Genetically-targeted therapy | ||
| Tazemetostat | EZH2 inhibitor | |
| Duvelisib | PI3K inhibitor | |
| Copanlisib | PI3K inhibitor | |
| Ibrutinib | CARD11 and FOXO1 mutation+ cases | |
| Vorinostat | CREBBP and EP300 mutation+ cases | |
| Pidilizumab | PD-L1 | |
Follicular lymphoma progresses into transformation to diffuse large B-cell lymphoma (associated to TP53 mutation); and high-grade B-cell lymphoma with double-hit BCL2 and MYC rearrangement in most cases.56 Nevertheless, transformation to Epstein–Barr virus-related Hodgkin-like lymphoma has been described.57 Other rarer forms also include classic Hodgkin lymphoma, histiocytic/dendritic sarcoma, and high-grade B-cell lymphoma with double-hit TdT+.56
B-cell leukemia/lymphoma 2 (BCL2)
Most follicular lymphoma cases overexpress the apoptosis regulator Bcl-2, also known as B-cell leukemia/lymphoma 2 (BCL2), which is located at the 18q21.33 chromosomal location. BCL2 overexpression in most cases is due to the translocation t(14;18)(q32;q21) that locates the BCL2 gene under the control of the IgH locus (85-90 percent of Follicular lymphoma cases). Other uncommon but equivalent translocations involving BCL2 are the kappa light chain gene on chromosome 2 resulting in t(2;18)(p11;q21); and the lambda light chain gene on chromosome 22 resulting in t(18;22)(q21;q21).58,59 In this rearrangement process, RAG1 and RAG2 are involved as they mediate breaks in pre-B cells.60
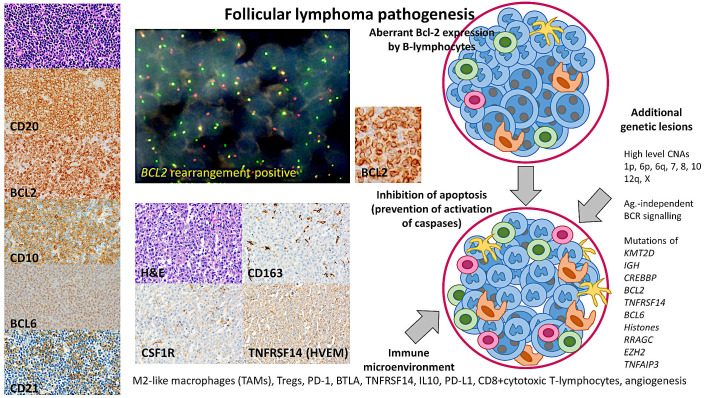
The BCL2 oncogene is a suppressor of apoptosis (prevents caspase activation) in many types of cells,61 and as a result the cells have increased survival. Nevertheless, the BCL2 overexpression is not sufficient for developing lymphoma as this translocation has been found in healthy individuals.62-64 In addition to BCL2, follicular lymphoma has increased expression of other antiapoptotic factors such as BCL-XL and MCL1; and lower expression of proapoptotic factors such as BAX and BAD. Both BAX and BAD are the major initiators of apoptosis; they aggregate into the mitochondrial membrane and release cytochrome c into the cytoplasm. As a result, there is an activation of caspase-9 and then caspase-3. Active caspase-3 is responsible for apoptotic chromatin condensation and DNA fragmentation.65-68 Therefore, the result is a promotion of cell survival69 (Figure 3).
Fig. 3.
Pathogenesis of follicular lymphoma
The pathogenesis of follicular lymphoma is multifactorial; and involves a series of steps in which a B lymphocyte acquires genetic and epigenetic alterations that lead to the malignant transformation. The pathobiology includes genetic alterations in BCL2 and BCL6, a mutational profile, and changes in the immune microenvironment and the immune checkpoint. On the left, a characteristic low-grade follicular lymphoma is shown. This case was characterized by a nodular proliferation of centrocytes, which had a classical phenotype with positivity of CD20, CD10, BCL6, and BCL2, and a mesh of CD21 follicular dendritic cells. CNAs, copy number alterations.
BCL2 rearrangement-negative follicular lymphoma represents less than 10% of the cases, and their protein expression of BCL2 is decreased or absent in comparison to the follicular lymphoma with BCL2 translocation.70 Usually, the rearrangement-negative follicular lymphoma cases are characterized by having a predominantly diffuse histological pattern; or a nodular/follicular growth pattern with a high histological grade (usually 3b). Interestingly, as described by Katzenberger T and Siddiqi IN et al.,71,72 these predominantly diffuse follicular lymphoma cases are atypical, have variable protein expression of BCL2, high protein expression of CD23, copy number losses of 1p36 locus, and mutations of TNF receptor superfamily member 14 (TNFRSF14) and STAT6 mutation.
BCL6 transcription repressor
Follicular lymphoma cases with a histological grade 3B appear closer to diffuse large B-cell lymphoma than low-grade follicular lymphoma.24 Cytogenetic studies found that could be characterized by t(14;18) affecting the BCL2 gene, the presence of 3q27 rearrangement involving the BCL6 gene, and others.24 Although initially it was thought that the BCL2 and BCL6 rearrangements were mutually exclusive. Nowadays, there are cases of double-positive rearrangement exist.73-76 B-cell lymphoma 6 protein (BCL6) is a transcriptional repressor that plays a role in the formation of the germinal center and in antibody affinity maturation. The mechanism of repression is by binding to the BCL6-binding site (5’-TTCCTAGAA-3’) or by repressing the transcriptional activity of other transcription factors.77 BCL6 is responsible for the suppression in B-lymphocytes of the germinal centers of genes associated with cell differentiation, apoptosis (TP53), cell cycle control (allowing them to proliferate), and inflammation. BCL6 upregulates Activation-induced cytidine deaminase (AICDA, also known as AID), which is responsible for germinal center-associated somatic hypermutation (SHM).32,77-83 BCL6-null mice lack germinal centers.84,85 Therefore, BCL6 is a key oncogene in B-cell lymphomagenesis.86
Other genetic alterations
Cytogenetic and whole-genome copy number and LOH analyses have shown that in follicular lymphoma, the most frequent areas of gains are located at chromosomes 1, 6p, 7, 8, 12q, X, and 18q/dup, and of losses of 1p, 6q, 10q, and 17p2 (Figure 3). Several genomic changes affect the 1p36 locus, which includes the TNFRSF14 gene, and include copy number losses, copy neutral loss of heterozygosity (CN-LOH, acquired uniparental disomy), and mutations. These changes are associated with poor prognosis.87-89 Deletions of 6q are found in around 20% of the cases, and these changes are usually associated with an unfavorable prognosis.88,90,91 The abnormalities of chromosome 3 include the 3q27 locus and involve the BCL6 gene; and are associated with BCL2 rearrangement-negative cases.24,73
MYC translocations are commonly acquired and are present in 25% of transformed follicular lymphoma cases.92,93 Recently, it has been reported that cases with BCL6 rearrangement and/or BCL6 gain (with cases of BCL2 rearrangement and/or of copy number gain excluded) correlated with favorable progression-free survival.75 BCL6 rearrangement-positive follicular lymphoma is characterized by higher rates of grade 3A, and MUM1 expression and less interfollicular spread pattern.75
Mutational landscape
The mutational landscape of follicular lymphoma has been analyzed by several groups using next-generation sequencing (NGS). The NGS workflow contains three steps: library preparation, sequencing, and analysis. The analysis tech-workflow includes primary, secondary, including mutation calling, and tertiary with annotation, and data interpretation and prioritization. Differences in highlighting high-confidence calls are a relevant issue to be solved.
Several groups have focused on the analysis of histone and chromatin-modifying genes. In order of frequency, the most mutated in follicular lymphoma were KMT2D, CREBBP, EZH2, EP300, HIST1H1E, KMT2C, ARID1A, and SMARCA4.93-101 Interestingly, it has been reported that CREBBP and EZH2 mutations are an early event in the pathogenesis, whereas KMT2D and TNFRSF14 would occur later.94,97,102
An important pathway that is also mutated is mTOR, such as RRAGC, which is important for the activation of the pathway.103 Interestingly, activating RRAGC mutations would promote lymphomagenesis by interacting with the microenvironment.104
Mutations of the B-cell receptor pathway are also found, including BTK; and CD79B, which control many functions of B-lymphocytes such as cell growth, differentiation, survival, and migration.105
Transformed follicular lymphoma is associated with TP53 mutations, and they are not usually found at diagnosis; but are found in subsequent biopsies before transformation.106-108
In order of frequency (Table 1), the most frequently mutated genes in follicular lymphoma are KMT2D (80-90%) as a loss of function (histone modification, proliferation); IgHV and IgLV, gain of function (75-90%, BCR signaling, proliferation); CREBBP (33-70%), loss of function (histone modification), BCL2 (50%), gain of function (suppression of apoptosis, survival); TNFRSF14 (20-50%), loss of function (increased BCR signaling, immune evasion); BCL6 (47%), gain of function (transcription factor, tumor progression); H1-2, H1-4 (44%), loss of function (chromatin remodeling); RRAGC (17%), gain of function (mTORC1 survival signal); EZH2 (7-30%), gain of function (histone modification), and TNFAIP3 (2-26%), loss of function (loss of tumor suppressor, survival)3 (Table 1) (Figure 3).
An improved prediction of follicular lymphoma using a targeted sequencing panel, known as m7-FLIPI, has also been designed. The calculator can be accessed at https://www.german-lymphoma-alliance.de/box.php?action=box.boilerplate.detail&site=scores&boilerplatePk=BD7B559B-C5CF-DF40-19B1-4E214D787FFA (Accessed on March 30, 2023). This panel included seven genes (EZH2, ARID1A, MEF2B, EP300, FOXO1, CREBBP and CARD11), FLIPI, and Eastern Cooperative Oncology Group (ECOG) performance status.109
The mutational profile of t(14;18) negative Follicular lymphoma has recently been described,53 and it has been found to be heterogeneous. The differences between stage I versus III/IV follicular lymphoma have also been analyzed.110
Finally, one of the few genetically targeted therapies available is tazemetostat. It is an inhibitor of EZH2 approved by the FDA that provided a favorable overall response rate in the case of EZH2 mutation.111
Immune checkpoint and tumor immune microenvironment
The knowledge of the composition and role of the tumor microenvironment (TME), host immune response, and immune checkpoint has recently advanced. Follicular lymphoma cells are mixed with a TME milieu of nonmalignant immune, stromal, and extracellular components, which create a bidirectional interaction between the follicular lymphoma neoplastic cells and the immune microenvironment.41,112,113
Since the identification of the role of the immune microenvironment in follicular lymphoma by gene expression arrays, many publications have analyzed several components of the microenvironment that were found to have prognostic relevance. The components highlighted are immune gene-signature, macrophages, CD4-positive T-lymphocytes, CD8-positive cytotoxic T-lymphocytes, FOXP-positive regulatory T-lymphocytes (Tregs), PD-1, and PD-1 ligands, TNFRSF14 (HVEM), BTLA, CSF1R, IL-10, and microvessel density, among others. In summary, the data indicate that markers of M2-like tumor-associated macrophages (TAMs) and microvessels (angiogenesis) are associated with an unfavorable prognosis. Conversely, FOXP3 (Tregs), PD-1 (Tfh), and CD8 (Tc) are associated with a favorable clinical evolution.29-49 Some components of the microenvironment are also related to the histological transformation, such as high levels of tumor-associated CD68-positive and PD-L1-positive macrophages (TAMs)114 (Figure 3 and 4).
Fig. 4.
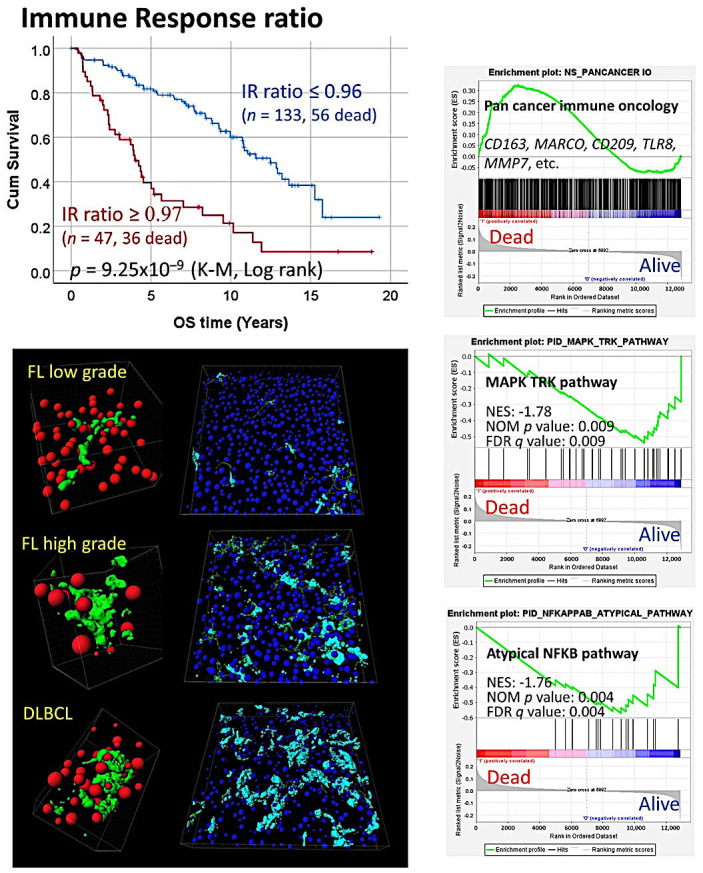
The immune response in follicular lymphoma
The immune response is central to the pathobiology of follicular lymphoma. The work of LLMPP36 identified two immune responses related to the host immune response with predictive value; the immune response type 2 was characterized by genes associated with tumor-associated macrophages. Further work using the same gene expression dataset confirmed the results, including several pathway analyses using gene set enrichment analysis (GSEA). Interestingly, macrophages increased in the progression from low- to high-grade follicular lymphoma but also in the transformation to diffuse large B-cell lymphoma, including the creation of 3D networks.34,35,142
A recent study analyzed the CSF-1/CSF1R pathway using an in vitro analysis and an in vivo follicular lymphoma xenograft mouse model. The results were validated in a series of human follicular lymphoma by immunohistochemistry. The crosstalk between follicular lymphoma B-lymphocytes, macrophages, and follicular dendritic cells was analyzed. This result supported the role of macrophages in follicular lymphoma pathogenesis; and indicated that CSF1R may be relevant as a prognostic factor cooperating with anti-CD20 immunotherapy.48
Precision medicine
The management of patients with relapse or refractory follicular lymphoma is changing. Cases with EZH2 mutations (in codon Y646, A682, or A692) with no alternative treatment options can be treated with tazemetostat, which is an antineoplastic agent and a potent and selective inhibitor of EZH2. Other drugs are duvelisib (PI3K inhibitor), umbralisib (multikinase inhibitors), and copanlisib (PI3K inhibitor). Nevertheless, FDA approval of the lymphoma medicine umbralisib has been withdrawn due to safety concerns.115,116
FDA has also granted accelerated approval to axicabtagene ciloleucel (Yescarta) for relapsed or refractory follicular lymphoma, which is an immunotherapy medicine (a CD19-directed chimeric antigen receptor (CAR) T-cell therapy).115,116
Other possible targets are ibrutinib (CARD11 and FOXO1 mutation), pidilizumab (PD-L1 expression), and vorinostat (CREBBP and EP300 mutation)117 (Table 1).
Pathobiology and highlights in the lymphoma classification
Recently, there have been advances in the understanding of the pathobiology of follicular lymphoma, which has led to changes and/or highlights in the classification of small B-cell lymphoid neoplasms of ICC 2022.5
• Histological grades are maintained as previously.2 In case of follicular lymphoma grade 3, the presence of rearrangement of BCL2 and CD10 positivity favor grade 3A against 3B. In young cases, grade 3B and MUM1 (IRF4) positivity should recommend analysis of IRF4 alteration. Routine molecular testing is not necessary; but could be recommended in selected patients (EZH2 inhibitors).
• BCL2-R negative; CD23-positive follicle center lymphoma is classified as a type of follicle center lymphoma. This type can have a histological diffuse pattern, is located in the pelvic/inguinal zone, and usually has STAT6 mutations.
• Primary cutaneous follicle center lymphoma has molecular and cytogenetic characteristics different from those of other follicular lymphoma.
• Testicular follicular lymphoma is recognized as a distinct form in young boys.
Advances in the pathobiology of Follicular lymphoma have also been translated into the WHO-HAEM5 as described by Alaggio R. et al..9
• The term classic follicular lymphoma (cFL) is used and is separated from follicular large B-cell lymphoma (FLBL); and follicular lymphoma with uncommon features (uFL). The classic FL is the most frequent (85%), has a follicular pattern, comprises centrocytes and centroblasts, and is characterized by the t(14;18) (q32;q21) translocation that is associated with the IGH::BCL2 fusion. Of note, in cFL, grading is no longer mandatory.
• Follicular lymphoma grade 3B equals FLBL.
• The uFL includes the “blastoid”; and the predominantly diffuse growth pattern (which corresponds to the BCL2 rearrangement-negative, CD23-positive follicle center cell lymphoma).
Artificial intelligence
The birth of artificial intelligence (AI) was denoted by Alan Turing’s seminal work “Computing Machinery and Intelligence”,118 which described AI as systems that act like humans. AI is the engineering of intelligent computer programs.119 AI combines computer science and robust datasets to solve problems.120 Using both machine learning and deep learning, it is possible to make predictions and classifications based on input data. A turning point in AI has been the release of OpenAI’s ChatGPT, which is a trained conversational model.121 Nevertheless, it is important to point out that thinking and making our own decisions is what makes us humans. Letting machines think for us makes us less free; and less conscious. Therefore, no machine should be made in the likeness of the human mind.
From 2015, there has been an exponential increase in the number of publications that use deep learning in the pathology field in Japan. Examples include gastrointestinal pathology,122 precision medicine,123 urothelial carcinoma,124 ocular pathology,125 esophageal cancer,126 lung cancer,127 thyroid cytology,128 intestinal diseases,122,129-135 sarcoma,136 hematological,137-140 among others. In the field of malignant lymphoma, Miyoshi H and Ohshima K et al. showed how deep learning was capable of high-level computer-aided diagnosis based on H&E slides, including diffuse large B-cell lymphoma, follicular lymphoma, and reactive lymphoid hyperplasia;141 and Hashimoto N and Takeuchi I et al. analyzed several malignant lymphoma cases using immunohistochemical patterns.138 In case of follicular lymphoma, the pathobiology has also been described using machine learning and neural networks, predicting the prognosis based on immune checkpoint and other oncogenes.34,35,142 In conclusion, the pathobiology of neoplasia and follicular lymphoma is being analyzed using new technology.
Conclusions
Follicular lymphoma is one of the most frequent lymphomas, and is a heterogeneous disease, both clinically and genetically. It is characterized by the t(14;18) with the IGH::BCL2 rearrangement, but additional genetic changes and changes in the microenvironment are necessary for the pathogenesis. The importance of other genetic changes as prognostic markers remains to be further developed.
ACKNOWLEDGMENTS
I want to thank Naoya Nakamura for revising the manuscript. This work was funded by grant Grant-in-Aid for Scientific Research KAKEN 23K06454, 18K15100, and 15K19061 from MEXT, Japan.
Footnotes
CONFLICT OF INTEREST
No conflicts of interest to declare.
REFERENCES
- 1.de Leval L, Jaffe ES. Lymphoma Classification. Cancer J. 2020; 26: 176-185. [DOI] [PubMed] [Google Scholar]
- 2.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016; 127: 2375-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carbone A, Roulland S, Gloghini A, et al. Follicular lymphoma. Nat Rev Dis Primers. 2019; 5: 83. [DOI] [PubMed] [Google Scholar]
- 4.Yoshino T, Takata K, Tanaka T, et al. Recent progress in follicular lymphoma in Japan and characteristics of the duodenal type. Pathol Int. 2018; 68: 665-676. [DOI] [PubMed] [Google Scholar]
- 5.Campo E, Jaffe ES, Cook JR, et al. The International Consensus Classification of Mature Lymphoid Neoplasms: a report from the Clinical Advisory Committee. Blood. 2022; 140(11): 1229-1253. Blood. 2023; 141: 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campo E, Jaffe ES, Cook JR, et al. The International Consensus Classification of Mature Lymphoid Neoplasms: a report from the Clinical Advisory Committee. Blood. 2022; 140: 1229-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cazzola M, Sehn LH. Developing a classification of hematologic neoplasms in the era of precision medicine. Blood. 2022; 140: 1193-1199. [DOI] [PubMed] [Google Scholar]
- 8.de Leval L, Alizadeh AA, Bergsagel PL, et al. Genomic profiling for clinical decision making in lymphoid neoplasms. Blood. 2022; 140: 2193-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alaggio R, Amador C, Anagnostopoulos I, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia. 2022; 36: 1720-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol. 2015; 15: 137-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaber T, Buttgereit F. A new perspective is needed for positive selection of germinal center B cells with higher-affinity B cell receptors. Cell Mol Immunol. 2022; 19: 145-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mesin L, Ersching J, Victora GD. Germinal Center B Cell Dynamics. Immunity. 2016; 45: 471-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stebegg M, Kumar SD, Silva-Cayetano A, et al. Regulation of the Germinal Center Response. Front Immunol. 2018; 9: 2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young C, Brink R. The unique biology of germinal center B cells. Immunity. 2021; 54: 1652-1664. [DOI] [PubMed] [Google Scholar]
- 15.Küppers R, Klein U, Hansmann ML, Rajewsky K. Cellular origin of human B-cell lymphomas. N Engl J Med. 1999; 341: 1520-1529. [DOI] [PubMed] [Google Scholar]
- 16.Wartenberg M, Vasil P, zum Bueschenfelde CM, et al. Somatic hypermutation analysis in follicular lymphoma provides evidence suggesting bidirectional cell migration between lymph node and bone marrow during disease progression and relapse. Haematologica. 2013; 98: 1433-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klien U, Goossens T, Fischer M, et al. Somatic hypermutation in normal and transformed human B cells. Immunol Rev. 1998; 162: 261-280. [DOI] [PubMed] [Google Scholar]
- 18.Halldórsdóttir AM, Frühwirth M, Deutsch A, et al. Quantifying the role of aberrant somatic hypermutation in transformation of follicular lymphoma. Leuk Res. 2008; 32: 1015-1021. [DOI] [PubMed] [Google Scholar]
- 19.Kosmas C, Stamatopoulos K, Papndoki T, et al. Somatic hypermutation of immunoglobulin variable region genes: focus on follicular lymphoma and multiple myeloma. Immunol Rev. 1998; 162: 281-292. [DOI] [PubMed] [Google Scholar]
- 20.Solal-Céligny P, Roy P, Colombat P, et al. Follicular lymphoma international prognostic index. Blood. 2004; 104: 1258-1265. [DOI] [PubMed] [Google Scholar]
- 21.Nooka AK, Nabhan C, Zhou X, et al. Examination of the follicular lymphoma international prognostic index (FLIPI) in the National LymphoCare study (NLCS): a prospective US patient cohort treated predominantly in community practices. Ann Oncol. 2013; 24: 441-448. [DOI] [PubMed] [Google Scholar]
- 22.Bachy E, Maurer MJ, Habermann TM, et al. A simplified scoring system in de novo follicular lymphoma treated initially with immunochemotherapy. Blood. 2018; 132: 49-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alig S, Jurinovic V, Pastore A, et al. Impact of age on clinical risk scores in follicular lymphoma. Blood Adv. 2019; 3: 1033-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bosga-Bouwer AG, van Imhoff GW, Boonstra R, et al. Follicular lymphoma grade 3B includes 3 cytogenetically defined subgroups with primary t(14;18), 3q27, or other translocations: t(14;18) and 3q27 are mutually exclusive. Blood. 2003; 101: 1149-1154. [DOI] [PubMed] [Google Scholar]
- 25.Harris NL, Jaffe ES, Diebold J, et al. World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the Clinical Advisory Committee meeting-Airlie House, Virginia, November 1997. J Clin Oncol. 1999; 17: 3835-3849. [DOI] [PubMed] [Google Scholar]
- 26.Horn H, Schmelter C, Leich E, et al. Follicular lymphoma grade 3B is a distinct neoplasm according to cytogenetic and immunohistochemical profiles. Haematologica. 2011; 96: 1327-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shustik J, Quinn M, Connors JM, et al. Follicular non-Hodgkin lymphoma grades 3A and 3B have a similar outcome and appear incurable with anthracycline-based therapy. Ann Oncol. 2011; 22: 1164-1169. [DOI] [PubMed] [Google Scholar]
- 28.Wahlin BE, Yri OE, Kimby E, et al. Clinical significance of the WHO grades of follicular lymphoma in a population-based cohort of 505 patients with long follow-up times. Br J Haematol. 2012; 156: 225-233. [DOI] [PubMed] [Google Scholar]
- 29.Álvaro T, Lejeune M, Salvadó MT, et al. Immunohistochemical patterns of reactive microenvironment are associated with clinicobiologic behavior in follicular lymphoma patients. J Clin Oncol. 2006; 24: 5350-5357. [DOI] [PubMed] [Google Scholar]
- 30.Canioni D, Salles G, Mounier N, et al. High numbers of tumor-associated macrophages have an adverse prognostic value that can be circumvented by rituximab in patients with follicular lymphoma enrolled onto the GELA-GOELAMS FL-2000 trial. J Clin Oncol. 2008; 26: 440-446. [DOI] [PubMed] [Google Scholar]
- 31.Carreras J, Lopez-Guillermo A, Fox BC, et al. High numbers of tumor-infiltrating FOXP3-positive regulatory T cells are associated with improved overall survival in follicular lymphoma. Blood. 2006; 108: 2957-2964. [DOI] [PubMed] [Google Scholar]
- 32.Carreras J, Lopez-Guillermo A, Kikuti YY, et al. High TNFRSF14 and low BTLA are associated with poor prognosis in Follicular Lymphoma and in Diffuse Large B-cell Lymphoma transformation. J Clin Exp Hematop. 2019; 59: 1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carreras J, Lopez-Guillermo A, Roncador G, et al. High numbers of tumor-infiltrating programmed cell death 1-positive regulatory lymphocytes are associated with improved overall survival in follicular lymphoma. J Clin Oncol. 2009; 27: 1470-1476. [DOI] [PubMed] [Google Scholar]
- 34.Carreras J, Kikuti YY, Miyaoka M, et al. Artificial Intelligence Analysis of the Gene Expression of Follicular Lymphoma Predicted the Overall Survival and Correlated with the Immune Microenvironment Response Signatures. Mach Learn Knowl Extr. 2020; 2: 647-671. [Google Scholar]
- 35.Carreras J, Kikuti YY, Miyaoka M, et al. The Use of the Random Number Generator and Artificial Intelligence Analysis for Dimensionality Reduction of Follicular Lymphoma Transcriptomic Data. BioMedInformatics. 2022; 2: 268-280. [Google Scholar]
- 36.Dave SS, Wright G, Tan B, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004; 351: 2159-2169. [DOI] [PubMed] [Google Scholar]
- 37.Farinha P, Kyle AH, Minchinton AI, et al. Vascularization predicts overall survival and risk of transformation in follicular lymphoma. Haematologica. 2010; 95: 2157-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farinha P, Masoudi H, Skinnider BF, et al. Analysis of multiple biomarkers shows that lymphoma-associated macrophage (LAM) content is an independent predictor of survival in follicular lymphoma (FL). Blood. 2005; 106: 2169-2174. [DOI] [PubMed] [Google Scholar]
- 39.Glas AM, Knoops L, Delahaye L, et al. Gene-expression and immunohistochemical study of specific T-cell subsets and accessory cell types in the transformation and prognosis of follicular lymphoma. J Clin Oncol. 2007; 25: 390-398. [DOI] [PubMed] [Google Scholar]
- 40.Klapper W, Hoster E, Rölver L, et al. Tumor sclerosis but not cell proliferation or malignancy grade is a prognostic marker in advanced-stage follicular lymphoma: the German Low Grade Lymphoma Study Group. J Clin Oncol. 2007; 25: 3330-3336. [DOI] [PubMed] [Google Scholar]
- 41.Kumar E, Pickard L, Okosun J. Pathogenesis of follicular lymphoma: genetics to the microenvironment to clinical translation. Br J Haematol. 2021; 194: 810-821. [DOI] [PubMed] [Google Scholar]
- 42.Küppers R. Prognosis in follicular lymphoma--it’s in the microenvironment. N Engl J Med. 2004; 351: 2152-2153. [DOI] [PubMed] [Google Scholar]
- 43.Lee AM, Clear AJ, Calaminici M, et al. Number of CD4+ cells and location of forkhead box protein P3-positive cells in diagnostic follicular lymphoma tissue microarrays correlates with outcome. J Clin Oncol. 2006; 24: 5052-5059. [DOI] [PubMed] [Google Scholar]
- 44.Tamma R, Ingravallo G, Annese T, et al. Tumor Microenvironment and Microvascular Density in Follicular Lymphoma. J Clin Med. 2022; 11: 1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taskinen M, Jantunen E, Kosma VM, et al. Prognostic impact of CD31-positive microvessel density in follicular lymphoma patients treated with immunochemotherapy. Eur J Cancer. 2010; 46: 2506-2512. [DOI] [PubMed] [Google Scholar]
- 46.Taskinen M, Karjalainen-Lindsberg ML, Nyman H, Eerola LM, Leppä S. A high tumor-associated macrophage content predicts favorable outcome in follicular lymphoma patients treated with rituximab and cyclophosphamide-doxorubicin-vincristine-prednisone. Clin Cancer Res. 2007; 13: 5784-5789. [DOI] [PubMed] [Google Scholar]
- 47.Tobin JWD, Keane C, Gunawardana J, et al. Progression of Disease Within 24 Months in Follicular Lymphoma Is Associated With Reduced Intratumoral Immune Infiltration. J Clin Oncol. 2019; 37: 3300-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valero JG, Matas-Céspedes A, Arenas F, et al. The receptor of the colony-stimulating factor-1 (CSF-1R) is a novel prognostic factor and therapeutic target in follicular lymphoma. Leukemia. 2021; 35: 2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wahlin BE, Sander B, Christensson B, Kimby E. CD8+ T-cell content in diagnostic lymph nodes measured by flow cytometry is a predictor of survival in follicular lymphoma. Clin Cancer Res. 2007; 13: 388-397. [DOI] [PubMed] [Google Scholar]
- 50.Louissaint A, Jr, Schafernak KT, Geyer JT, et al. Pediatric-type nodal follicular lymphoma: a biologically distinct lymphoma with frequent MAPK pathway mutations. Blood. 2016; 128: 1093-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt J, Ramis-Zaldivar JE, Nadeu F, et al. Mutations of MAP2K1 are frequent in pediatric-type follicular lymphoma and result in ERK pathway activation. Blood. 2017; 130: 323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt J, Gong S, Marafioti T, et al. Genome-wide analysis of pediatric-type follicular lymphoma reveals low genetic complexity and recurrent alterations of TNFRSF14 gene. Blood. 2016; 128: 1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nann D, Ramis-Zaldivar JE, Müller I, et al. Follicular lymphoma t(14;18)-negative is genetically a heterogeneous disease. Blood Adv. 2020; 4: 5652-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.JC. BJFAA. Pathobiology of follicular lymphoma. UpToDate. 2023.
- 55.Loeffler-Wirth H, Kreuz M, Schmidt M, et al. Classifying Germinal Center Derived Lymphomas-Navigate a Complex Transcriptional Landscape. Cancers (Basel). 2022; 14: 3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaffe ES, Carbone A. Evolution in the Definition of Follicular Lymphoma and Diffuse Large B-Cell Lymphoma: A Model for the Future of Personalized Medicine. Hemato. 2022; 3: 466-474. [Google Scholar]
- 57.Menon MP, Hutchinson L, Garver J, Jaffe ES, Woda BA. Transformation of follicular lymphoma to Epstein-Barr virus-related Hodgkin-like lymphoma. J Clin Oncol. 2013; 31: e53-e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bloomfield CD, Arthur DC, Frizzera G, et al. Nonrandom chromosome abnormalities in lymphoma. Cancer Res. 1983; 43: 2975-2984. [PubMed] [Google Scholar]
- 59.Rowley JD. Chromosome studies in the non-Hodgkin’s lymphomas: the role of the 14;18 translocation. J Clin Oncol. 1988; 6: 919-925. [DOI] [PubMed] [Google Scholar]
- 60.Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004; 428: 88-93. [DOI] [PubMed] [Google Scholar]
- 61.Eguchi Y, Ewert DL, Tsujimoto Y. Isolation and characterization of the chicken bcl-2 gene: expression in a variety of tissues including lymphoid and neuronal organs in adult and embryo. Nucleic Acids Res. 1992; 20: 4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levy D, Bertoldi ERM, Ruiz JLM, Pereira J, Bydlowski SP. Presence of t(14;18) translocation in healthy individuals varies according to ethnic background in the Brazilian population. Braz J Med Biol Res. 2017; 50: e6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roulland S, Lebailly P, Lecluse Y, et al. Long-term clonal persistence and evolution of t(14;18)-bearing B cells in healthy individuals. Leukemia. 2006; 20: 158-162. [DOI] [PubMed] [Google Scholar]
- 64.Schüler F, Hirt C, Dölken G. Chromosomal translocation t(14;18) in healthy individuals. Semin Cancer Biol. 2003; 13: 203-209. [DOI] [PubMed] [Google Scholar]
- 65.Porter AG, Jänicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999; 6: 99-104. [DOI] [PubMed] [Google Scholar]
- 66.Cory S, Adams JM. Killing cancer cells by flipping the Bcl-2/Bax switch. Cancer Cell. 2005; 8: 5-6. [DOI] [PubMed] [Google Scholar]
- 67.Boise LH, González-García M, Postema CE, et al. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993; 74: 597-608. [DOI] [PubMed] [Google Scholar]
- 68.Oltval ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programed cell death. Cell. 1993; 74: 609-619. [DOI] [PubMed] [Google Scholar]
- 69.Ghia P, Boussiotis VA, Schultze JL, et al. Unbalanced expression of bcl-2 family proteins in follicular lymphoma: contribution of CD40 signaling in promoting survival. Blood. 1998; 91: 244-251. [PubMed] [Google Scholar]
- 70.Schraders M, de Jong D, Kluin P, Groenen P, van Krieken H. Lack of Bcl-2 expression in follicular lymphoma may be caused by mutations in theBCL2 gene or by absence of the t(14;18) translocation. J Pathol. 2005; 205: 329-335. [DOI] [PubMed] [Google Scholar]
- 71.Katzenberger T, Kalla J, Leich E, et al. A distinctive subtype of t(14;18)-negative nodal follicular non-Hodgkin lymphoma characterized by a predominantly diffuse growth pattern and deletions in the chromosomal region 1p36. Blood. 2009; 113: 1053-1061. [DOI] [PubMed] [Google Scholar]
- 72.Siddiqi IN, Friedman J, Barry-Holson KQ, et al. Characterization of a variant of t(14;18) negative nodal diffuse follicular lymphoma with CD23 expression, 1p36/TNFRSF14 abnormalities, and STAT6 mutations. Mod Pathol. 2016; 29: 570-581. [DOI] [PubMed] [Google Scholar]
- 73.Díaz-Alderete A, Doval A, Camacho F, et al. Frequency of BCL2 and BCL6 translocations in follicular lymphoma: relation with histological and clinical features. Leuk Lymphoma. 2008; 49: 95-101. [DOI] [PubMed] [Google Scholar]
- 74.Miyaoka M, Kikuti YY, Carreras J, et al. Clinicopathological and genomic analysis of double-hit follicular lymphoma: comparison with high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements. Mod Pathol. 2018; 31: 313-326. [DOI] [PubMed] [Google Scholar]
- 75.Ikoma H, Miyaoka M, Hiraiwa S, et al. Clinicopathological analysis of follicular lymphoma with BCL2, BCL6, and MYC rearrangements. Pathol Int. 2022; 72: 321-331. [DOI] [PubMed] [Google Scholar]
- 76.Takeoka K, Maekawa F, Nakagawa M, et al. MYC/BCL2 double- and MYC/BCL2/BCL6 triple-hit follicular lymphomas associated with t(8;14;18)(q24;q32;q21). J Clin Exp Hematop. 2022; 62: 258-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ci W, Polo JM, Cerchietti L, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009; 113: 5536-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leung W, Teater M, Durmaz C, et al. SETD2 Haploinsufficiency Enhances Germinal Center-Associated AICDA Somatic Hypermutation to Drive B-cell Lymphomagenesis. Cancer Discov. 2022; 12: 1782-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007; 76: 1-22. [DOI] [PubMed] [Google Scholar]
- 80.Teater M, Dominguez PM, Redmond D, et al. AICDA drives epigenetic heterogeneity and accelerates germinal center-derived lymphomagenesis. Nat Commun. 2018; 9: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fairfax KA, Gantier MP, Mackay F, Williams BRG, McCoy CE. IL-10 regulates Aicda expression through miR-155. J Leukoc Biol. 2015; 97: 71-78. [DOI] [PubMed] [Google Scholar]
- 82.Zan H, Casali P. Regulation of Aicda expression and AID activity. Autoimmunity. 2013; 46: 83-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miyaoka M, Kikuti YY, Carreras J, et al. AID is a poor prognostic marker of high‐grade B‐cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements. Pathol Int. 2022; 72: 35-42. [DOI] [PubMed] [Google Scholar]
- 84.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997; 276: 589-592. [DOI] [PubMed] [Google Scholar]
- 85.Ye BH, Cattoretti G, Shen Q, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997; 16: 161-170. [DOI] [PubMed] [Google Scholar]
- 86.Basso K, Dalla-Favera R. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv Immunol. 2010; 105: 193-210. [DOI] [PubMed] [Google Scholar]
- 87.Cheung KJ, Johnson NA, Affleck JG, et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res. 2010; 70: 9166-9174. [DOI] [PubMed] [Google Scholar]
- 88.Cheung KJ, Shah SP, Steidl C, et al. Genome-wide profiling of follicular lymphoma by array comparative genomic hybridization reveals prognostically significant DNA copy number imbalances. Blood. 2009; 113: 137-148. [DOI] [PubMed] [Google Scholar]
- 89.Launay E, Pangault C, Bertrand P, et al. High rate of TNFRSF14 gene alterations related to 1p36 region in de novo follicular lymphoma and impact on prognosis. Leukemia. 2012; 26: 559-562. [DOI] [PubMed] [Google Scholar]
- 90.Gaidano G, Ballerini P, Gong JZ, et al. p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 1991; 88: 5413-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schwaenen C, Viardot A, Berger H, et al. Microarray-based genomic profiling reveals novel genomic aberrations in follicular lymphoma which associate with patient survival and gene expression status. Genes Chromosomes Cancer. 2009; 48: 39-54. [DOI] [PubMed] [Google Scholar]
- 92.Kridel R, Mottok A, Farinha P, et al. Cell of origin of transformed follicular lymphoma. Blood. 2015; 126: 2118-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pasqualucci L, Khiabanian H, Fangazio M, et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014; 6: 130-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Green MR, Gentles AJ, Nair RV, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013; 121: 1604-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pasqualucci L, Dominguez-Sola D, Chiarenza A, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011; 471: 189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011; 476: 298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bödör C, Grossmann V, Popov N, et al. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood. 2013; 122: 3165-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Okosun J, Bödör C, Wang J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014; 46: 176-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Green MR, Kihira S, Liu CL, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci USA. 2015; 112: E1116-E1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kridel R, Chan FC, Mottok A, et al. Histological Transformation and Progression in Follicular Lymphoma: A Clonal Evolution Study. PLoS Med. 2016; 13: e1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Green MR. Chromatin modifying gene mutations in follicular lymphoma. Blood. 2018; 131: 595-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schmidt J, Ramis-Zaldivar JE, Bonzheim I, et al. CREBBP gene mutations are frequently detected in in situ follicular neoplasia. Blood. 2018; 132: 2687-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Okosun J, Wolfson RL, Wang J, et al. Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma. Nat Genet. 2016; 48: 183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ortega-Molina A, Deleyto-Seldas N, Carreras J, et al. Oncogenic Rag GTPase signalling enhances B cell activation and drives follicular lymphoma sensitive to pharmacological inhibition of mTOR. Nat Metab. 2019; 1: 775-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Krysiak K, Gomez F, White BS, et al. Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma. Blood. 2017; 129: 473-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Davies AJ, Lee AM, Taylor C, et al. A limited role for TP53 mutation in the transformation of follicular lymphoma to diffuse large B-cell lymphoma. Leukemia. 2005; 19: 1459-1465. [DOI] [PubMed] [Google Scholar]
- 107.Lo Coco F, Gaidano G, Louie DC, et al. p53 mutations are associated with histologic transformation of follicular lymphoma. Blood. 1993; 82: 2289-2295. [PubMed] [Google Scholar]
- 108.Sander CA, Yano T, Clark HM, et al. p53 mutation is associated with progression in follicular lymphomas. Blood. 1993; 82: 1994-2004. [PubMed] [Google Scholar]
- 109.Pastore A, Jurinovic V, Kridel R, et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol. 2015; 16: 1111-1122. [DOI] [PubMed] [Google Scholar]
- 110.Los-de Vries GT, Stevens WBC, van Dijk E, et al. Genomic and microenvironmental landscape of stage I follicular lymphoma, compared with stage III/IV. Blood Adv. 2022; 6: 5482-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Morschhauser F, Tilly H, Chaidos A, et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: an open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol. 2020; 21: 1433-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dobaño-López C, Araujo-Ayala F, Serrat N, Valero JG, Pérez-Galán P. Follicular Lymphoma Microenvironment: An Intricate Network Ready for Therapeutic Intervention. Cancers (Basel). 2021; 13: 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.López C, Mozas P, López-Guillermo A, Beà S. Molecular Pathogenesis of Follicular Lymphoma: From Genetics to Clinical Practice. Hemato. 2022; 3: 595-614. [Google Scholar]
- 114.Blaker YN, Spetalen S, Brodtkorb M, et al. The tumour microenvironment influences survival and time to transformation in follicular lymphoma in the rituximab era. Br J Haematol. 2016; 175: 102-114. [DOI] [PubMed] [Google Scholar]
- 115.U.S. Food & Drug Administration (FDA) 2023 [Available from: https://www.fda.gov/.
- 116.Drugs.com . 2023. [Available from: https://www.drugs.com/.
- 117.Sorigue M, Cañamero E, Sancho JM. Precision medicine in follicular lymphoma: focus on predictive biomarkers. Hematol Oncol. 2020; 38: 625-639. [DOI] [PubMed] [Google Scholar]
- 118.Turing AM. I.—COMPUTING MACHINERY AND INTELLIGENCE. Mind. 1950; LIX: 433-460. [Google Scholar]
- 119.McCarthy J. WHAT IS ARTIFICIAL INTELLIGENCE? Computer Science Department, Stanford University. 2007. [Google Scholar]
- 120.Russell S, Norvig P. Artificial Intelligence: A Modern Approach, 4th US ed. Aug 22, 2022 ed2022 Aug 22, 2022.
- 121.Open AI. Introducing ChatGPT 2023 [Available from: https://openai.com/blog/chatgpt.
- 122.Yoshida H, Kiyuna T. Requirements for implementation of artificial intelligence in the practice of gastrointestinal pathology. World J Gastroenterol. 2021; 27: 2818-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hamamoto R, Suvarna K, Yamada M, et al. Application of Artificial Intelligence Technology in Oncology: Towards the Establishment of Precision Medicine. Cancers (Basel). 2020; 12: 3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nojima S, Terayama K, Shimoura S, et al. A deep learning system to diagnose the malignant potential of urothelial carcinoma cells in cytology specimens. Cancer Cytopathol. 2021; 129: 984-995. [DOI] [PubMed] [Google Scholar]
- 125.Maruyama K, Mei S, Sakaguchi H, et al. Diagnosis of Choroidal Disease With Deep Learning-Based Image Enhancement and Volumetric Quantification of Optical Coherence Tomography. Transl Vis Sci Technol. 2022; 11: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Horie Y, Yoshio T, Aoyama K, et al. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019; 89: 25-32. [DOI] [PubMed] [Google Scholar]
- 127.Kobayashi K, Bolatkan A, Shiina S, Hamamoto R. Fully-Connected Neural Networks with Reduced Parameterization for Predicting Histological Types of Lung Cancer from Somatic Mutations. Biomolecules. 2020; 10: 1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hirokawa M, Niioka H, Suzuki A, et al. Application of deep learning as an ancillary diagnostic tool for thyroid FNA cytology. Cancer Cytopathol. 2023; 131: 217-225. [DOI] [PubMed] [Google Scholar]
- 129.Yamamoto Y, Carreras J, Shimizu T, et al. Anti-HBV drug entecavir ameliorates DSS-induced colitis through PD-L1 induction. Pharmacol Res. 2022; 179: 105918. [DOI] [PubMed] [Google Scholar]
- 130.Takenaka K, Kawamoto A, Okamoto R, Watanabe M, Ohtsuka K. Artificial intelligence for endoscopy in inflammatory bowel disease. Intest Res. 2022; 20: 165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stidham RW, Takenaka K. Artificial Intelligence for Disease Assessment in Inflammatory Bowel Disease: How Will it Change Our Practice? Gastroenterology. 2022; 162: 1493-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ohara J, Nemoto T, Maeda Y, et al. Deep learning-based automated quantification of goblet cell mucus using histological images as a predictor of clinical relapse of ulcerative colitis with endoscopic remission. J Gastroenterol. 2022; 57: 962-970. [DOI] [PubMed] [Google Scholar]
- 133.Noguchi T, Ando T, Emoto S, et al. Artificial Intelligence Program to Predict p53 Mutations in Ulcerative Colitis-Associated Cancer or Dysplasia. Inflamm Bowel Dis. 2022; 28: 1072-1080. [DOI] [PubMed] [Google Scholar]
- 134.Carreras J. Artificial Intelligence Analysis of Celiac Disease Using an Autoimmune Discovery Transcriptomic Panel Highlighted Pathogenic Genes including BTLA. Healthcare (Basel). 2022; 10: 1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Carreras J. Artificial Intelligence Analysis of Ulcerative Colitis Using an Autoimmune Discovery Transcriptomic Panel. Healthcare (Basel). 2022; 10: 1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Maeda Y, Watanabe T, Izumi T, et al. Biomolecular Fluorescence Complementation Profiling and Artificial Intelligence Structure Prediction of the Kaposi’s Sarcoma-Associated Herpesvirus ORF18 and ORF30 Interaction. Int J Mol Sci. 2022; 23: 9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tomita H, Yamashiro T, Iida G, et al. Unenhanced CT texture analysis with machine learning for differentiating between nasopharyngeal cancer and nasopharyngeal malignant lymphoma. Nagoya J Med Sci. 2021; 83: 135-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hashimoto N, Takagi Y, Masuda H, et al. Case-based similar image retrieval for weakly annotated large histopathological images of malignant lymphoma using deep metric learning. Med Image Anal. 2023; 85: 102752. [DOI] [PubMed] [Google Scholar]
- 139.Carreras J, Nakamura N, Hamoudi R. Artificial Intelligence Analysis of Gene Expression Predicted the Overall Survival of Mantle Cell Lymphoma and a Large Pan-Cancer Series. Healthcare (Basel). 2022; 10: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Carreras J, Hamoudi R, Nakamura N. Artificial Intelligence Analysis of Gene Expression Data Predicted the Prognosis of Patients with Diffuse Large B-Cell Lymphoma. Tokai J Exp Clin Med. 2020; 45: 37-48. [PubMed] [Google Scholar]
- 141.Miyoshi H, Sato K, Kabeya Y, et al. Deep learning shows the capability of high-level computer-aided diagnosis in malignant lymphoma. Lab Invest. 2020; 100: 1300-1310. [DOI] [PubMed] [Google Scholar]
- 142.Carreras J, Roncador G, Hamoudi R. Artificial Intelligence Predicted Overall Survival and Classified Mature B-Cell Neoplasms Based on Immuno-Oncology and Immune Checkpoint Panels. Cancers (Basel). 2022; 14: 5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Küppers R, Stevenson FK. Critical influences on the pathogenesis of follicular lymphoma. Blood. 2018; 131: 2297-2306. [DOI] [PubMed] [Google Scholar]