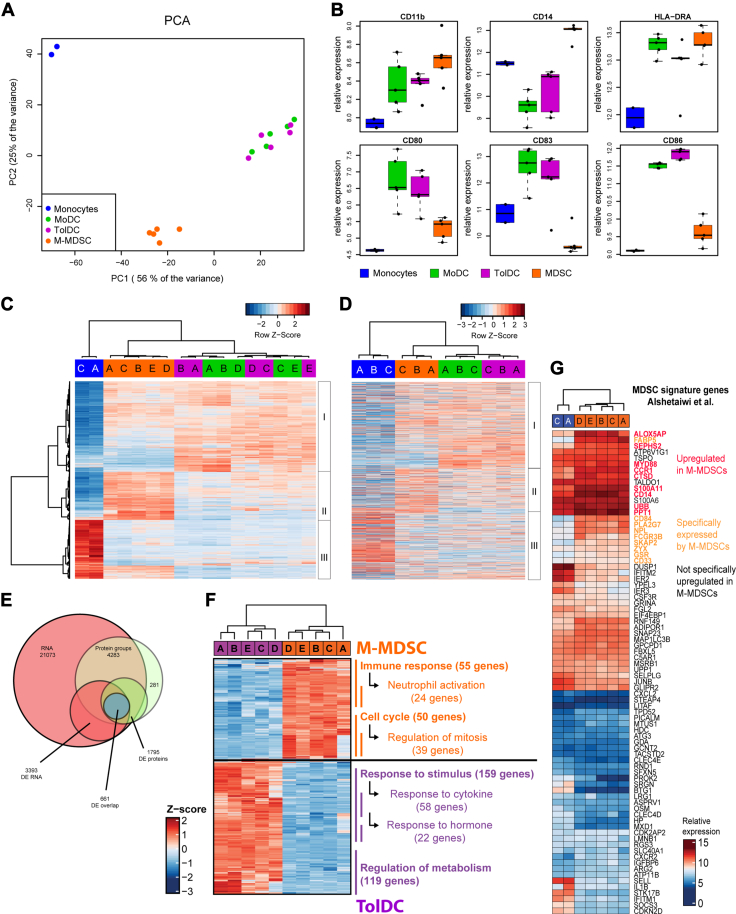
Figure 3.
Transcriptome and proteome analysis reveals clusters of differentially expressed genes and proteins between generated M-MDSCs and TolDCs. Generated myeloid cells (n = 5 donors) were assessed by Affymetrix HGU-133 Plus 2.0 RNA microarray and shown are (A) principal component analysis of top1000 variably expressed probes between subsets, (B) boxplots of genes encoding phenotypic markers, (C) heatmap of all significantly (p < 0.05) and differentially expressed (log2FC > 2) probes between all samples which identifies three clusters of differentially expressed genes (DEGs). Colors indicate cell types, and letters indicate individual donors. D, heatmap showing expression of proteins corresponding to the DEG in panel C as detected by mass spectrometry of generated myeloid cells (n = 3 donors). The same three differentially expressed clusters are also identified. E, Venn diagram showing overlap between the gene and protein expression data including DEG and DEP. F, Geneset enrichment analysis using Gene Ontology terms identifies the biological processes that are differentially activated between M-MDSCs and TolDCs. G, heatmap showing the expression of genes from a previously proposed MDSC signature by Alshetaiwi et al. among the human monocytes and generated M-MDSCs used in this study. Mean + SD in panel B. DEP, differentially expressed protein; MDSC, myeloid-derived suppressor cell; M-MDSC, monocyte-derived-MDSC; TolDC, tolerogenic dendritic cell.