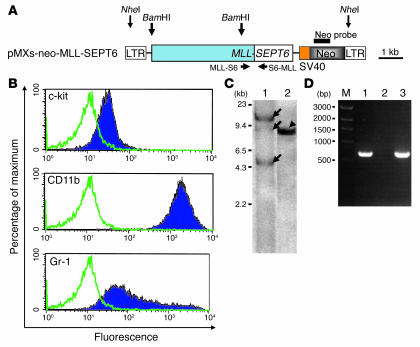
Figure 2.
Characterization of the cells immortalized by MLL-SEPT6. (A) Schematic representation of pMXs-neo-MLL-SEPT6. The restriction endonuclease sites and the Neo probe used in the Southern blot analysis are indicated by vertical arrows and a thick horizontal line, respectively. The primers used are indicated by horizontal arrows. LTR, long-terminal repeat. (B) Immunophenotype of the cells immortalized by MLL-SEPT6. Shadow profiles represent FACS staining obtained with Abs specific for the indicated cell surface antigens. Green lines represent staining obtained with isotype control Abs. (C) Southern blot analysis to detect clonality (lane 1) and integration (lane 2). Genomic DNA extracted from the cells immortalized by MLL-SEPT6 was digested with BamHI (lane 1) or NheI (lane 2), and hybridized with the Neo probe. Oligoclonal bands of proviral integration sites and a single band of the intact proviral DNA are indicated by arrows and an arrowhead, respectively. (D) Expression of MLL-SEPT6 fusion transcripts by RT-PCR. M, 1-kb DNA ladder (New England Biolabs Inc.); lane 1, cells immortalized by MLL-SEPT6; lane 2, negative control; lane 3, positive control (pMXs-neo-MLL-SEPT6).