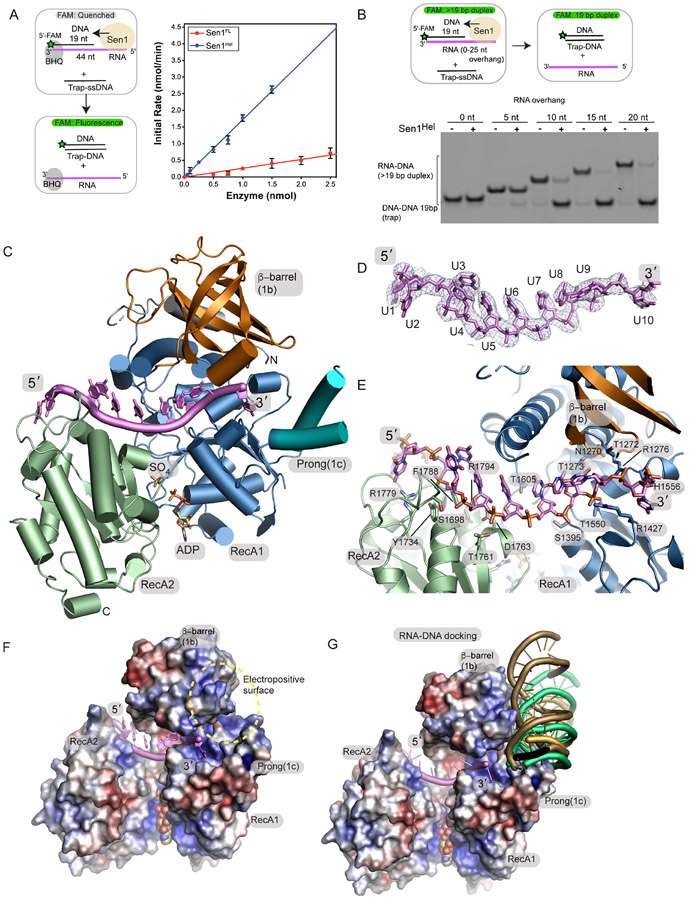
Figure 2. Crystal structure of the Sen1Hel–ssRNA–ADP–SO4 complex.
(A) Left: RNA-DNA unwinding substrates. Right: Sen1FL and Sen1Hel at the amounts indicated were incubated with 50 nmol of Sub7 (Table S2). Reactions were initiated by addition of a mixture containing ATP (1 mM) and MgCl2 (2 mM). FAM signal was detected at 520 nm every 10 sec over 30 min at 25 °C. Initial rates were determined by linear fit of the first 5 min of the unwinding reaction. Error bars are SD from 5 replicates.
(B) Top: gel-based RNA-DNA unwinding assay. Bottom: Effect of RNA overhang length on Sen1Hel RNA-DNA unwinding activity. Sub1-Sub5 (Supplementary Table 3) at 20 nM, Sen1Hel (50 nM), 19-Trap DNA (Table 1) at 200 nM, ATP (1 mM), MgCl2 (2 mM) were incubated for 15 min at 37 °C.
(C) X-ray structure of the Sen1Hel–RNA–ADP–SO4 complex. The extended LZZ coiled coil is not shown.
(D) Composite Omit 2Fo-Fc electron density at 1.0 σ, carved 2.0 Å around the RNA chain.
(E) Sen1Hel RNA-protein interactions show an extensive RNA binding interface.
(F) APBS surface electrostatic surface representation (blue=electropositive, red= electronegative) of the Sen1 RNA binding surface. Sen1 encircles the RNA via its RecA1 and RecA2 domains.
(G) Representative HADDOCK RNA-DNA poses displayed show secondary putative RNA-DNA hybrid binding electropositive surface flanks the ssRNA binding site and is assembled by the 1b and 1c RecA1 insertions.