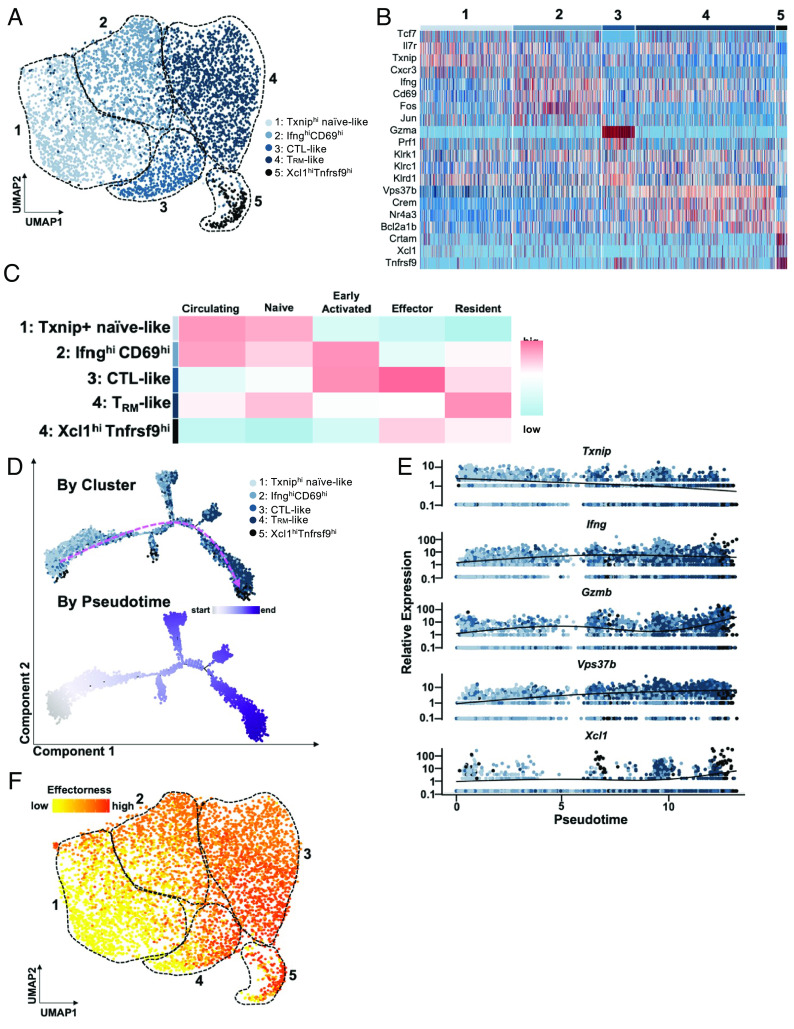
Fig. 3.
An effectorness gradient underlies CD8+ T cell heterogeneity in AA skin and identifies a potential trajectory for their differentiation. (A) UMAP of AA CD8+ T cell subsets. Unsupervised re-clustering of AA CD8+ T cells uncovered five subpopulations. (B) Heatmap of top highly expressed genes in each cluster. Cluster 1 showed higher expression of Tcf7, Il7r, Txnip, and Cxcr3; cluster 2 showed upregulation of Ifng, Cd69, Fos, and Jun; cluster 3 was enriched for Gzma, Prf1, Klrc1, and Klrd1; cluster 4 was up-regulated for Vps37b, Crem, Nr4a3, Bcl2a1b; cluster 5 showed higher expression of Crtam, Xcl1, and Tnfrsf9. Interestingly, expression of Klrk1, which encodes NKG2D, was not restricted to a single cluster. (C) Heatmap of scores for signatures of CD8+ T cell subpopulations described in previously published studies. Note that expression of gene signatures varies across clusters on a gradual continuum, as opposed to being exclusive to a particular cluster. (D) Pseudotime trajectory analysis of AA CD8+ T cells, colored by cluster (Top) and by position along pseudotime (Bottom). Pink dashed arrow denotes general direction of inferred differentiation across pseudotime. (E) Expression of select marker genes as identified in Fig. 3B along pseudotime, colored by cluster. (F) Pseudotime values were used to compute effectorness scores for each cell. Effectorness scores overlaid onto the UMAP of AA-associated CD8+ T cells demonstrated a high degree of correlation with the separation of the five CD8+ T cell subsets via unsupervised clustering of their transcriptional profiles, as shown in (A).