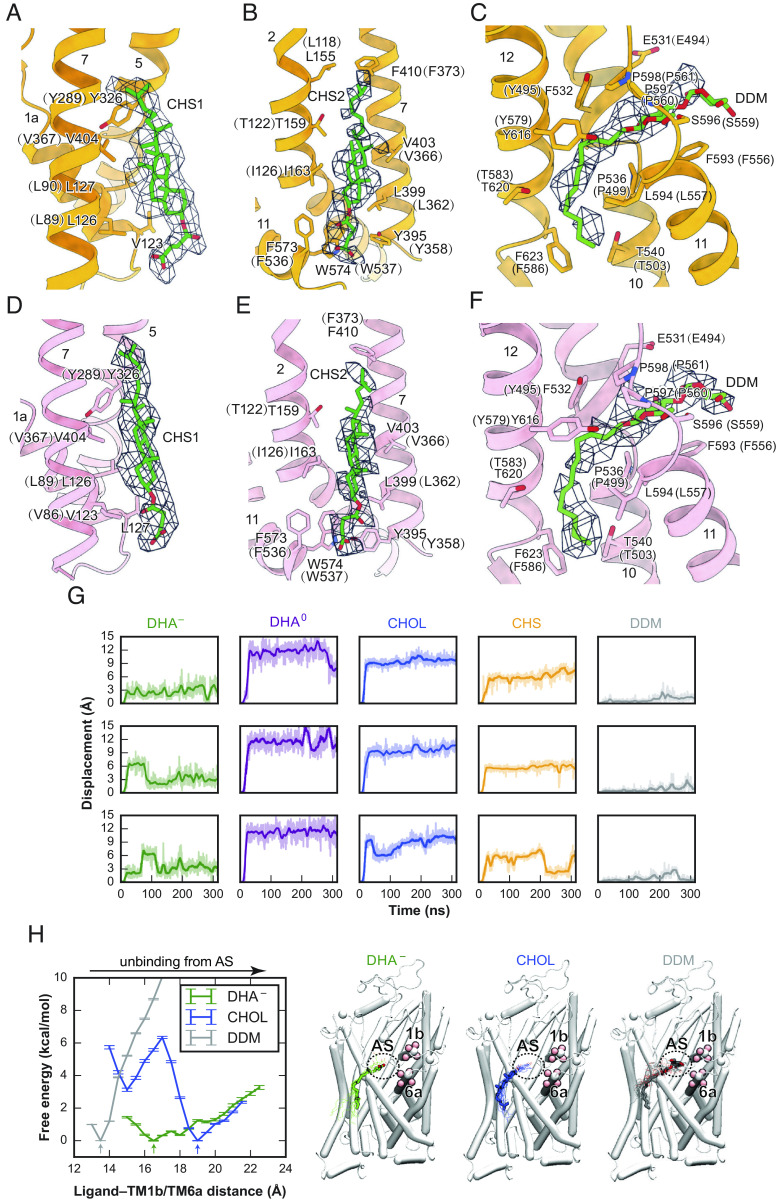
Fig. 4.
Cholesteryl hemisuccinate (CHS) and DDM binding in pSERT. (A and D) Close-up views of CHS modeled at the junction of TM1, TM5, and TM7 interacting with multiple hydrophobic residues. (B and E) CHS modeled at the junction of TM2, TM7, and TM11. (C and F) DDM modeled at the allosteric site. Shown are residue numbers for pSERT and in parenthesis, for hSERT, in panels A–F. (G) Time series of center-of-mass displacements of the ligands at the allosteric site (AS). Data for DHA–, DHA0, CHOL, CHS, and DDM are plotted in green, purple, blue, orange, and gray, respectively, and are shown for the three independent simulations in each case. Plots are smoothed using a sliding window of 1 ns. (H) Free energy profiles of DHA–, CHOL, and DDM binding to the allosteric site measured along the ligand–TM1b/TM6a distance, with molecular images showing each molecule in its most energetically favorable position (arrowed in left image). The ligand–TM1b/TM6a distance is measured as the center-of-mass distance between heavy atoms in the ligand and Cα atoms from TM1b and TM6a (residues 145 to 148 and 361 to 364, shown as pink spheres). The licorice representations represent DHA–, CHOL, and DDM in their most energetically favorable positions among all the examined ones (shown in line representations).