Figure 2: Prioritizing candidate causal variants at disease-associated loci.
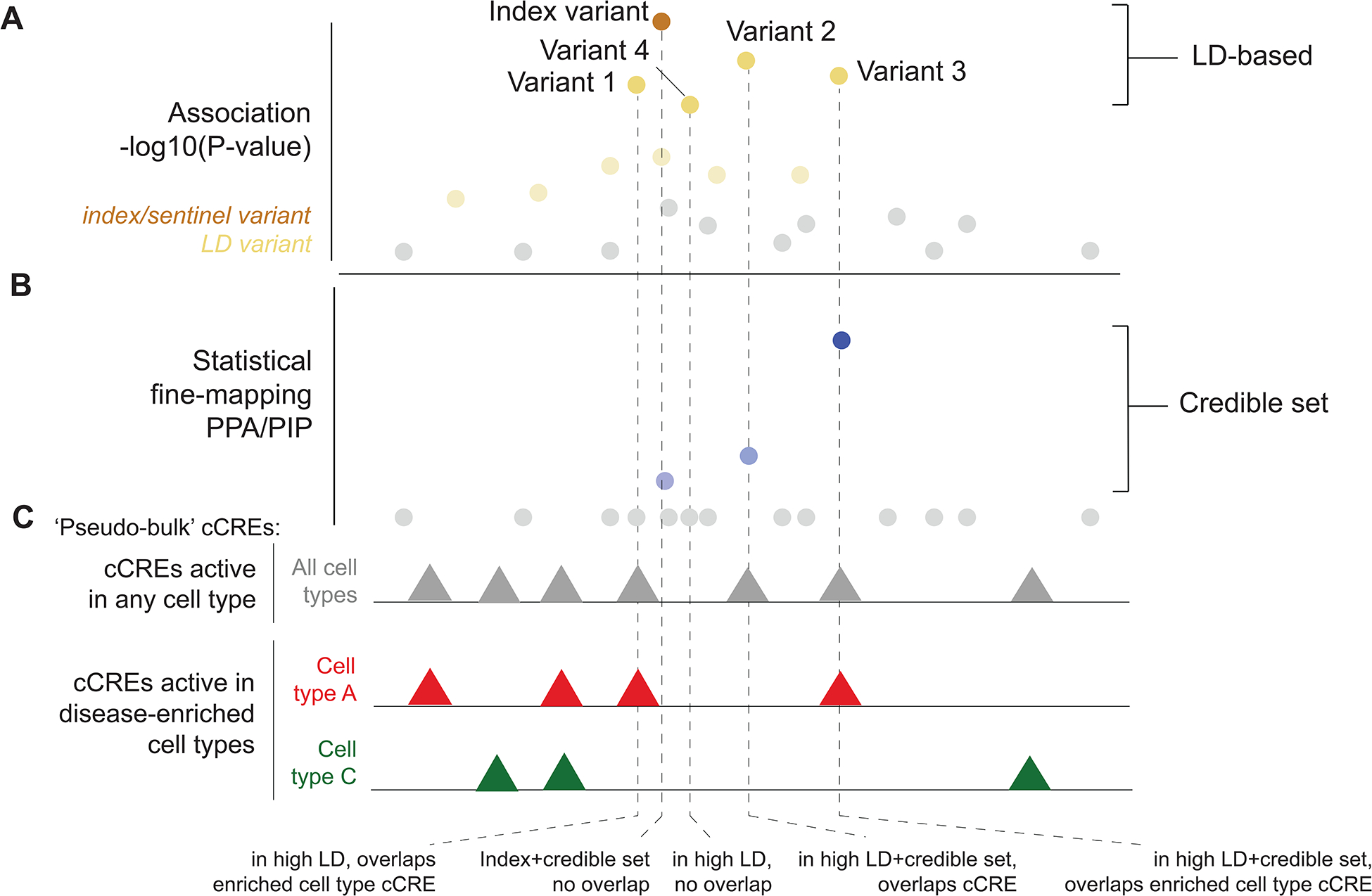
Overview of approaches for prioritizing variants causal for disease-associated loci used in single-cell epigenomics studies. a, Variant association data at a locus, where the most strongly associated ‘index’ variant and variants in linkage disequilibrium (LD) with the index variant are highlighted. In an LD-based approach, variants in high LD (typically r2>.8, although not specified in this example) with the index variant are prioritized as causal candidates, in this case variants 1–4. b, Statistical fine mapping derives the posterior probability that variants at a locus are causal for a risk signal. In a ‘credible set’ approach, variants that cumulatively explain a certain amount of the posterior probability (for example, 99% or 95%) of a risk signal from fine-mapping are prioritized as candidates. In this example, the index variant and variants 3 and 4 are in the credible set and thus prioritized by fine-mapping. c, cCREs identified using pseudo-bulk profiles of each cell type mapping to a locus are used to further prioritize among candidate variants. Studies have considered either cCREs active in any cell type, or just cCREs active in disease-enriched cell types. When prioritizing variants based on overlap with cCREs (indicated by dashed lines), variants 1, 3 and 4 overlap cCREs active in any cell type and variant 1 and 4 overlap cCREs active in a disease-enriched cell type. In this example, the index variant itself does not overlap any cCREs. Note that in these examples the predicted mechanism is that the causal variant affects cCRE activity, but in reality, variants can affect other regulatory mechanisms such as RNA splicing or stability.
