Figure 4: Linking disease variants to putative target genes.
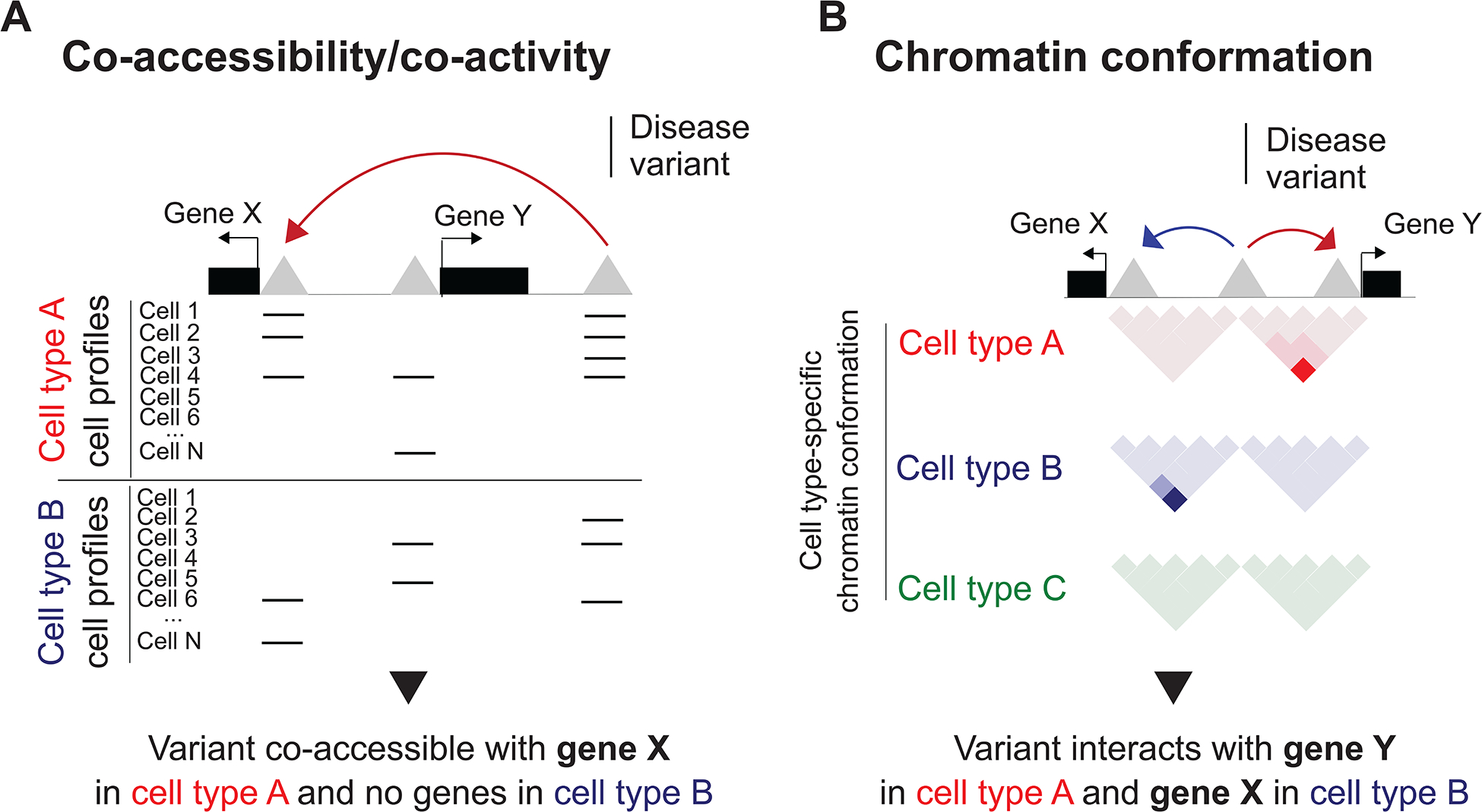
Overview of approaches for assigning disease variants to their target genes in specific cell types using single-cell epigenomics. a, Co-accessibility or co-activity of distal cCREs and promoter cCREs (both represented by grey triangles) across single-cells within a cell type using single-cell accessible chromatin data. In this example, a distal cCRE harboring a disease variant is accessible (represented by horizontal lines) in the same cells as the promoter cCRE of gene X but not gene Y in cell type A, indicating that gene X is the target gene of the distal cCRE in this cell type. By contrast, the distal cCRE is not co-accessible with either of the promoter cCREs in cell type B, suggesting it does not regulate either gene X or gene Y in this cell type b, Chromatin contacts between distal cCREs and promoter cCREs in specific cell types using single-cell 3D chromatin interaction data. In this example, a distal cCRE harboring a disease variant physically interacts with the promoter of gene Y in cell type A and with the promoter of gene X in cell type B. In cell type C, the distal cCRE does not interact with either of the genes.
