Figure 6. Characterization of ieAM and formation of ieAM organoids.
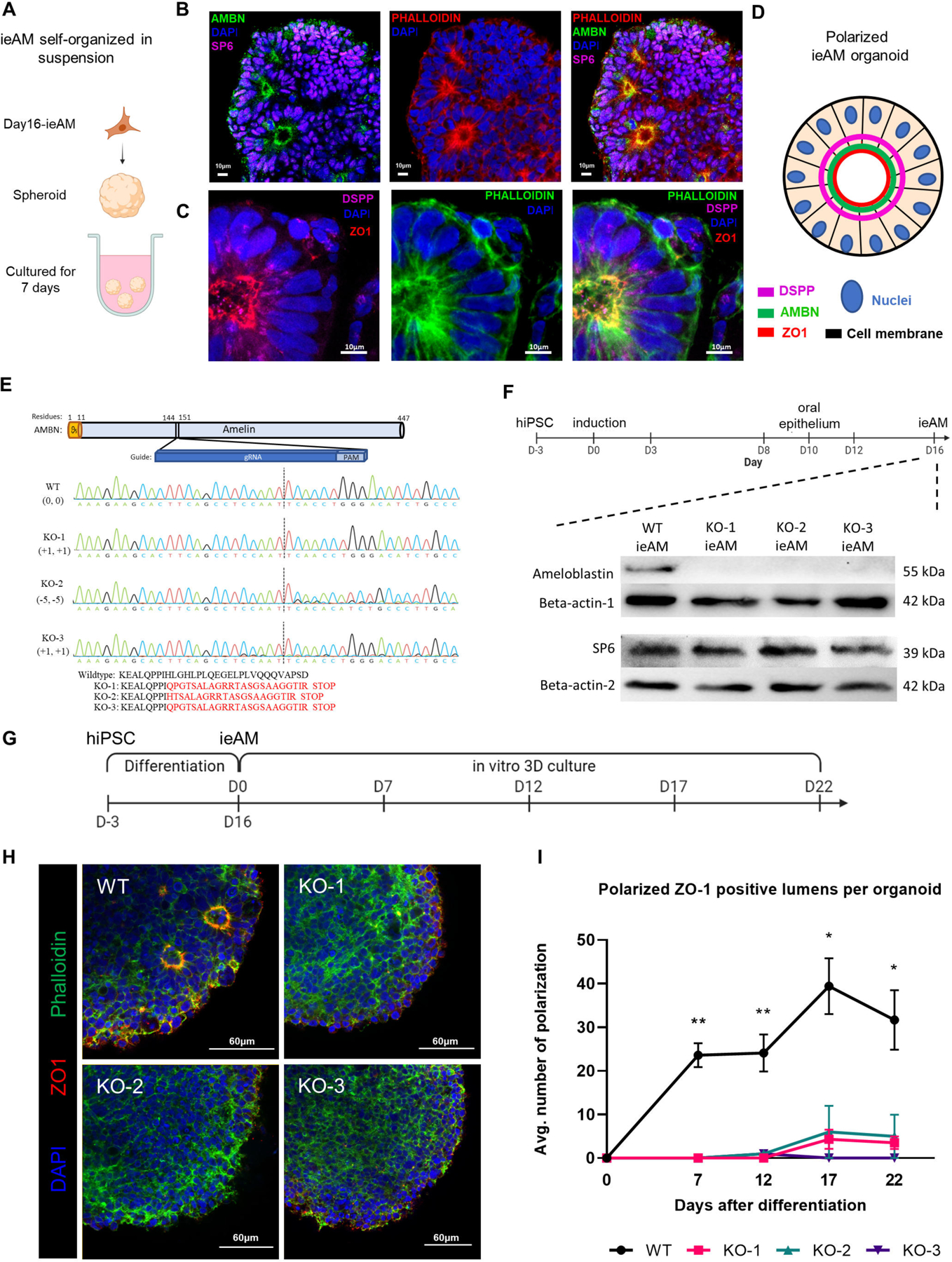
(A) Schematic of iAM organoids formation while cultured in suspension in ultra-low attachment plate. The formed iAM organoids express SP6 in the nuclei and AMBN (B), DSPP and ZO-1 (C) toward the apical side of the polarized ameloblasts. (D) A diagram simplifying the ameloblast organoid polarized structure toward a central lumen marked by ZO1, DSPP and AMBN. The markers observed indicate that the ameloblasts are in early stage of development (ieAM; high expression of AMBN and low expression of DSPP). (E) Protein structure of ameloblastin with guide RNA location indicated, as well as DNA sequencing chromatograms to compare the wild-type AMBN with the three AMBN mutant DNA sequences. The protein sequences below show that the three mutant cell lines have an early stop codon in their AMBN gene. KO-2 and KO-3 have a small population (<5%) of +1 and −11 respectively. (F) Western blot analysis showing AMBN protein knocked out. SP6 protein is a transcription factor of the AMBN gene, which acts as a marker for early-ameloblasts. The timeline of ieAM differentiation is shown above, indicating that the Western blot analysis was done on day 16. (G) Timeline of the ieAM differentiation plus in vitro 3D organoids culturing. (H) Representative confocal microscopy images showing cross-sections of in vitro 3D-cultured organoids derived from wild-type and mutant early-ameloblast lines stained with DAPI (blue), Phalloidin (F-actin, green) and ZO-1 (tight junction protein 1, red). Scale: 60μm. (I): Quantification of polarized ZO-1 positive lumens among in vitro 3D-cultured wild-type and mutant organoids at various time points after the 16-day ameloblast differentiation. *p ≤ 0.05 and ** p ≤ 0.01
