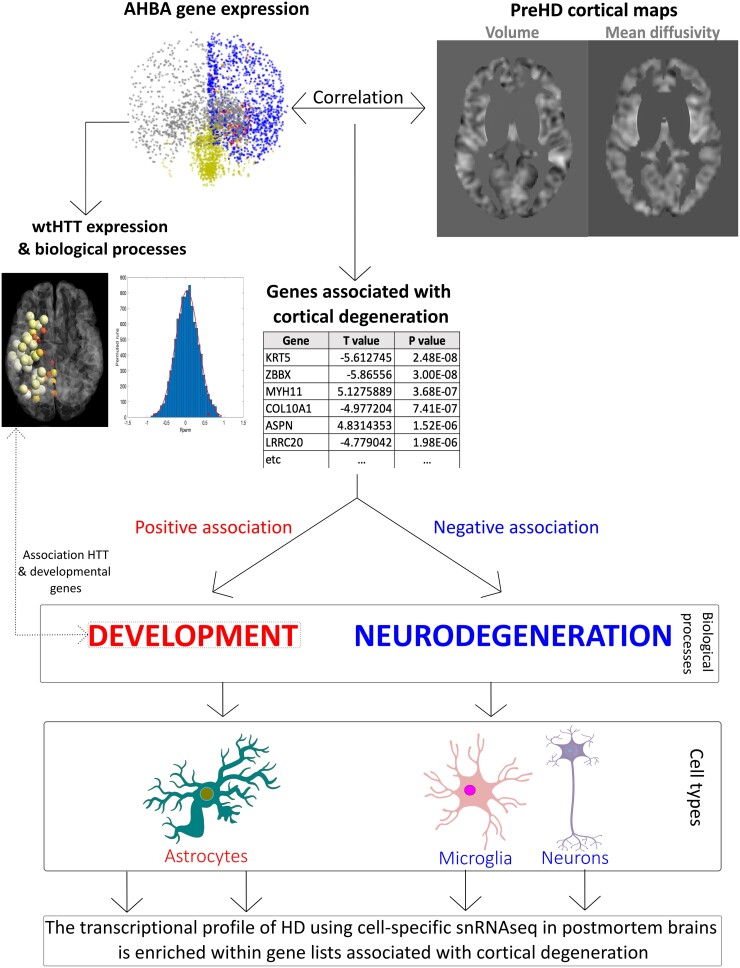
Figure 1.
Summary of analysis pipeline and main results. T-maps examining areas with lower cortical volume and higher mean diffusivity (cortical cell loss) in premanifest Huntington’s disease (preHD) compared to healthy controls were obtained and correlated with Allen Human Brain Atlas (AHBA) data at the voxel-level. Gene lists positively and negatively associated with cortical cell loss in preHD were provided. Genes positively associated with cortical cell loss were involved in development and enriched in transcriptomic profiles from astrocytes. The expression of HTT in the healthy human brain was associated with developmental biological processes and correlated with the expression of developmental genes. Genes negatively associated with cortical cell loss were involved in neurodegenerative (synaptic and metabolic) biological processes, being enriched in transcriptional profiles from neurons and microglial cells. The cell-specific transcriptional profiles from post-mortem Huntington’s disease (HD) brains using single-nucleus RNA sequencing (snRNAseq) were enriched within gene lists associated with cortical cell loss, affecting the same cell types and in the same directions as the ones found in the EWCE analyses. wt = wild-type.