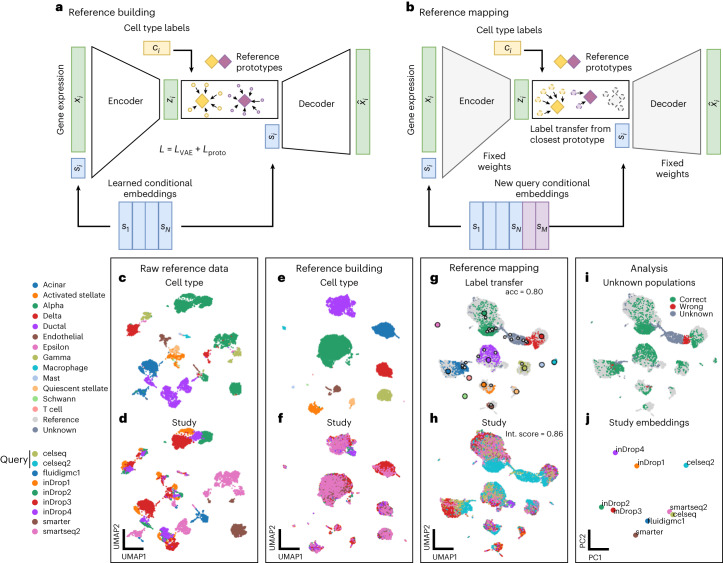
Fig. 1. scPoli enables learning cell-level and sample-level representations.
a, scPoli reference building: the model integrates different datasets and learns condition embeddings for each integrated study and a set of cell type prototypes. b, scPoli reference mapping: the model weights are frozen (in gray) and a new set of condition embeddings are added to the model. Cell type labels are transferred from the closest prototype in the latent space. Example of a standard workflow using scPoli on multiple pancreas datasets. c,d, Uniform manifold approximation and projection (UMAP) of the raw data to be integrated in a reference (13,093 cells), showing cell types (c) and studies (d) by color. e,f, Integrated reference data colored by cell type (e) and study (f). g, A total of 3,289 query cells (celseq and celseq2 studies) are projected onto the reference data in the reference mapping step. UMAPs show in color the query cells and in gray the reference cells. Reference cell type prototypes are shown in bigger circles with a black edge. Unlabeled prototypes are shown in bigger gray circles with black edges. The accuracy of the label transfer is 80%. h, Cells are colored by study or origin after reference mapping. The model achieves a mean integration score of 0.86. i, Outcome of the label transfer step from reference to query. j, PCA of the condition embeddings learned by scPoli.