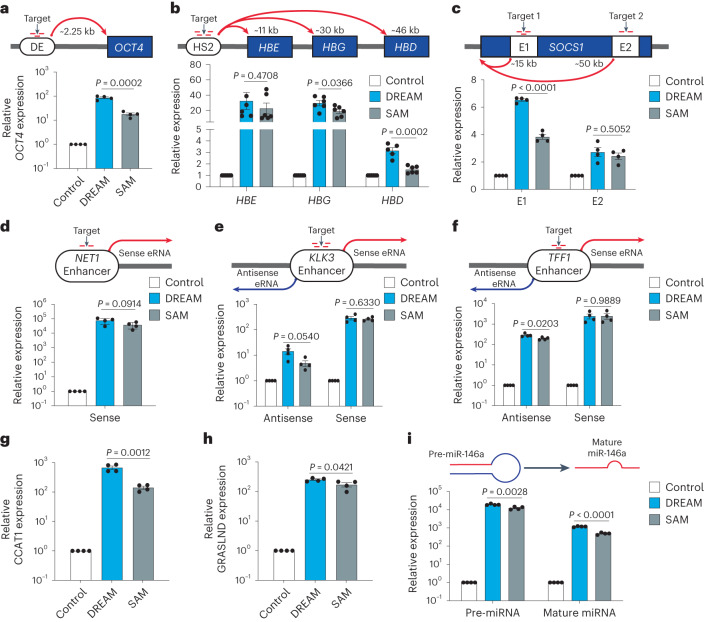
Fig. 2. CRISPR-DREAM efficiently activates transcription from diverse human regulatory elements.
a–c, CRISPR-DREAM and the SAM system activated downstream mRNA expression from OCT4 (a); HBE, HBG and HBD (b); and SOCS1 (c), when targeted to the OCT4 distal enhancer (DE), HS2 enhancer or one of two intragenic SOCS1 enhancers, using pools of 3 (OCT4 DE), 4 (HS2), 3 (SOCS1 + 15 kb) or 2 (SOCS1 + 50 kb) gRNAs, respectively. d, CRISPR-DREAM and the SAM system activated sense eRNA expression when targeted to the NET1 enhancer using 2 gRNAs. e,f, CRISPR-DREAM and the SAM system bidirectionally activated eRNA expression when targeted to the KLK3 (e) or TFF1 (f) enhancers using pools of 4 or 3 gRNAs, respectively. g,h, CRISPR-DREAM and the SAM system activated the expression of lncRNA when targeted to the CCAT1 (g) or GRASLND (h) promoters using pools of 4 gRNAs, respectively. i, CRISPR-DREAM and the SAM system activated the expression of pre- and mature miR-146a when targeted to the miR-146a promoter using a pool of 4 gRNAs. All samples were processed for qPCR at 72 h post-transfection. Data are the result of 5 or 6 biological replicates for b and 4 biological replicates for a and c–i. See the source data for more information. Data are presented as mean ± s.e.m. P values were determined using unpaired two-sided t-test. E1, Enhancer 1; E2, Enhancer 2.