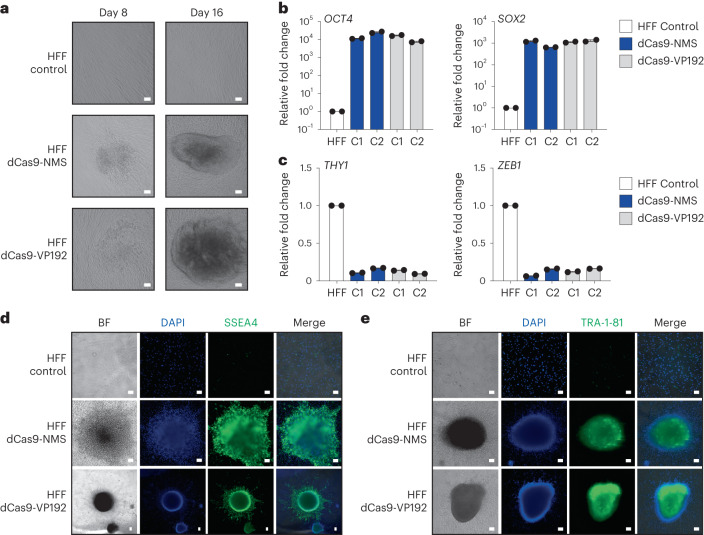
Fig. 5. dCas9-NMS permits efficient in vitro reprogramming of human fibroblasts.
a, Primary HFFs were nucleofected with plasmids encoding 15 multiplexed gRNAs targeting the OCT4, SOX2, KLF4, c-MYC and LIN28A promoter and EEA motifs (as previously reported17), and either dCas9-NMS (middle row) or dCas9-VP192 (bottom row). HFF morphology was analyzed 8 and 16 d later (white scale bars, 100 μm). b, Relative expression of pluripotency-associated genes OCT4 (left) and SOX2 (right) in representative iPSC colonies (C1 or C2) approximately 40 d after nucleofection of either dCas9-NMS (blue) or dCas9-VP192 (gray) and multiplexed gRNAs compared with untreated HFF controls. n = 2 independent measurements from independent subclones per colony. c, Relative expression of mesenchymal-associated genes THY1 (left) and ZEB1 (right) in representative iPSC colonies (C1 or C2) approximately 40 d after nucleofection of either dCas9-NMS (blue) or dCas9-VP192 (gray) and multiplexed gRNAs compared with untreated HFF controls. n = 2 independent measurements from independent subclones per colony. d,e, Immunofluorescence microscopy of HFFs approximately 40 d after nucleofection of either dCas9-NMS or dCas9-VP192 and multiplexed gRNAs compared with untreated HFF controls (white scale bars, 100 μm). Cells were stained for the expression of pluripotency-associated cell surface markers SSEA4 (d, green) or TRA-1-81 (e, green). All cells were counterstained with DAPI for nuclear visualization. Data presented in a is a representative of 3 independent experiments. Data are the result of 2 biological independent measurements from independent subclones per colony for b and c. Data presented in d and e are representative of 2 independent experiments. Data are presented as mean ± s.e.m.