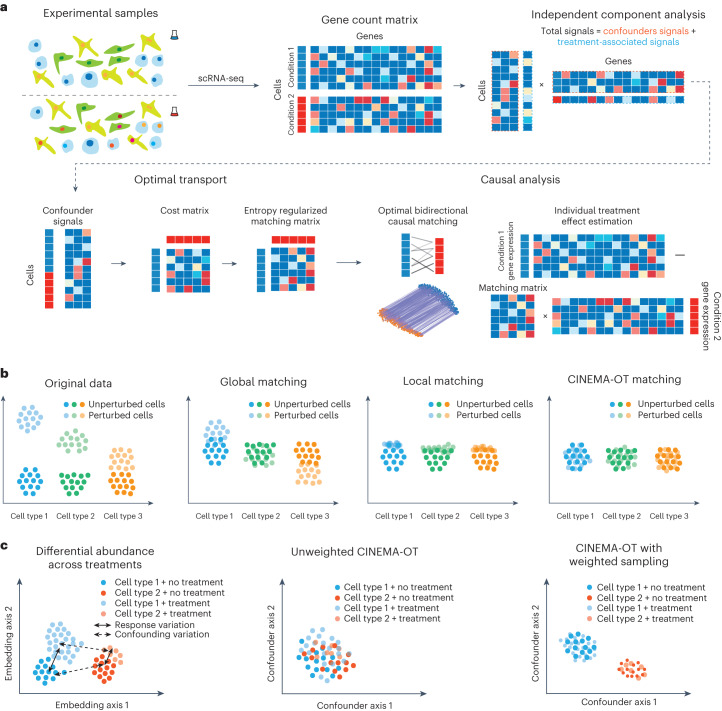
Fig. 1. Overview of the CINEMA-OT framework.
a, scRNA-seq count data is first decomposed into confounder variation and treatment-associated variation using ICA. Cells are then matched across treatment conditions by entropy-regularized optimal transport in the confounder space to generate a causal matching plan. The smooth matching map can then be used to estimate individual treatment effects. b, Illustration of the properties of CINEMA-OT compared with other potential matching schemes, including global matching (minimizing the average difference) and local matching (finding nearest neighbors for each cell). c, Illustration of the differential abundance issue in the unweighted CINEMA-OT method, and the resampling procedure used in CINEMA-OT-W.