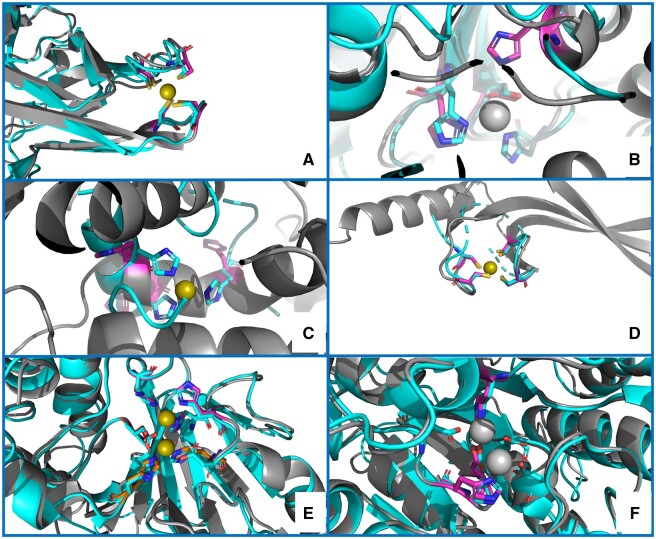
Figure 2.
Examples of structure predictions by MOM. (A) a correct prediction, validated by superimposition to a distant homolog structure [PDB ID 5ZLQ (Furukawa et al. 2018)]; (B) a partial match, with two out of three residues correctly superimposed to the metal ligands of the manganese(II) ion of a distant homolog structure [PDB ID 5M45 (Mus et al. 2017)]; (C) an inaccurate prediction [superimposed to the 1BM6_2 (Li et al. 1998) MetalPDB site], in which two of the three predicted His have a plausible spatial disposition, but the third His cannot be regarded a putative ligand since its positioning in the α-helix prevents any movement to form an MBS in the presence of the metal ion; (D) a correct prediction for a protein lacking a homolog with known structure, validated by superposition of the AlphaFold structural model to the closest MetalPDB site [3BVO_1 (Bitto et al. 2008)] identified by MoM; (E) a correct prediction for two zinc(II) sites in spatial proximity, validated by superimposition to a homolog structure [PDB ID 2P18 (Sousa Silva et al. 2008)], which contains a dinuclear zinc(II) cluster; (F) a partial match, where MoM predicted only one site containing a subset of the ligands to the two manganese(II) ions present in a homolog structure (1WVB). The color code is as follows: gray, AlphaFold structural models; fuchsia, predicted ligand residues; cyan, homolog structures or closest MetalPDB site. The zinc(II) ions are shown as olive green spheres, whereas all other metal ions are shown as grey spheres