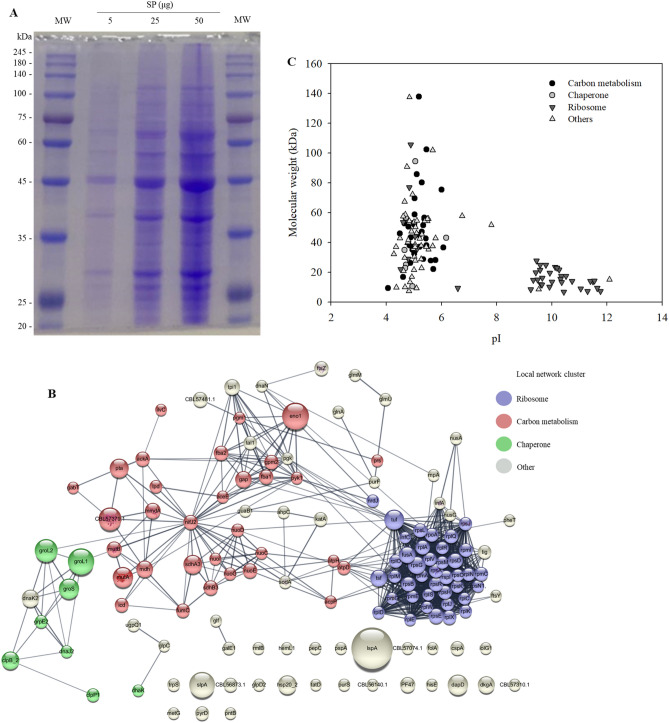
Figure 2.
Characterization of surface proteins (SP) isolated from Propionibacterium freudenreichii MJ2 using 5 M guanidine hydrochloride. (A) SP (5‒50 μg) were separated by electrophoresis on 10% sodium dodecyl sulfate–polyacrylamide gel. The full-length SDS-PAGE image of SP is shown in Supplementary Fig. 3. (B) STRING analysis of interaction networks among SP. Proteins are presented as nodes connected by edges. The size of nodes represents the protein score analyzed using the MASCOT algorithm and the color of the node indicates the local network cluster defined using STRING. The interactions of proteins were assessed under a high confidence cutoff (0.7). Purple, red, and green nodes indicate the network clusters related to ribosome, carbon metabolism, and chaperone, respectively. The thickness of edges indicates the confidence level of protein interaction score. (C) Molecular weight and isoelectric point (pI) of the identified proteins.