Figure 5. HIF2α inhibition cooperates with ATRA to promote AML differentiation.
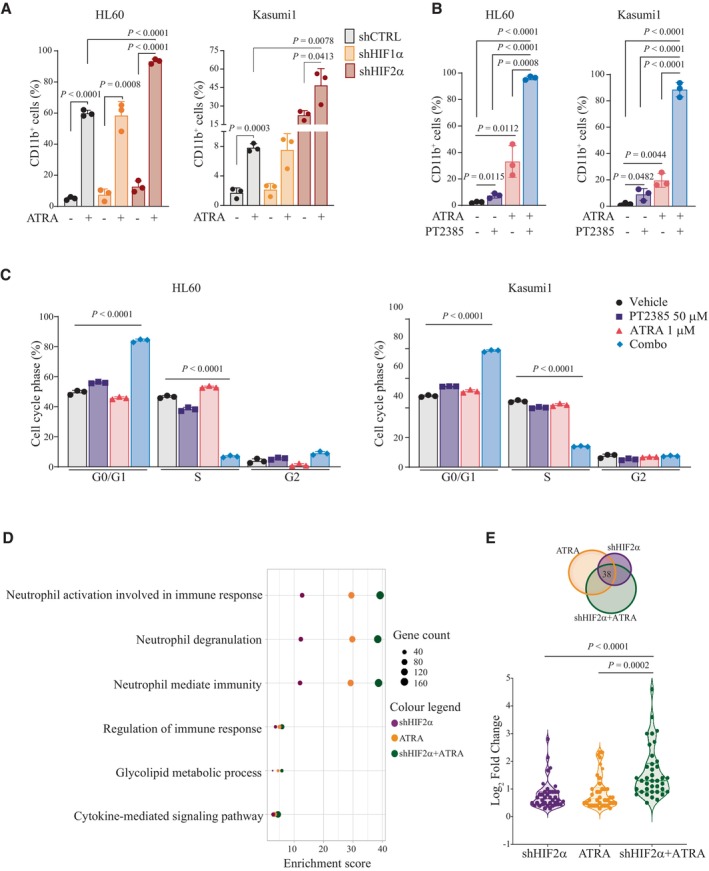
- Percentages of CD11b+ HL60 and Kasumi1 cells with shRNAs against HIF1α, HIF2α, or a scrambled shRNA as control (shCTRL) treated with 1 μM ATRA for 2 days. Data represent mean ± SD of three biological replicates (one‐way ANOVA followed by Tukey's multiple comparison test).
- Percentages of CD11b+ HL60 and Kasumi1 cells following treatment with 50 μM PT2385, 1 μM ATRA, or combination for 4 days. Data represent mean ± SD of three biological replicates (one‐way ANOVA followed by Tukey's multiple comparison test).
- Percentages of HL60 (left graph) and Kasumi1 (right graph) cells in the indicated phases of the cell cycle following treatment with 50 μM PT2385, 1 μM ATRA, or combination for 4 days. Data represent mean ± SD of three biological replicates (Student's t‐test).
- List of top common upregulated Gene Ontology (GO) terms in Kasumi1 cells upon shHIF2α, 1 μM ATRA treatment or combination with respect to shCTRL cells. Dot sizes represent the number of genes in each term, and colors indicate experimental conditions shown in legend.
- Venn diagram indicating the overlap of commonly upregulated genes (38 genes), which are represented in terms shown in (D). Violin plot indicating fold induction of each of the 38 genes commonly upregulated in each condition. Values represent the Log2 (FoldChange) with respect to shCTRL cells. Data indicate fold enrichment over control cells (Student's t‐test).
Source data are available online for this figure.
