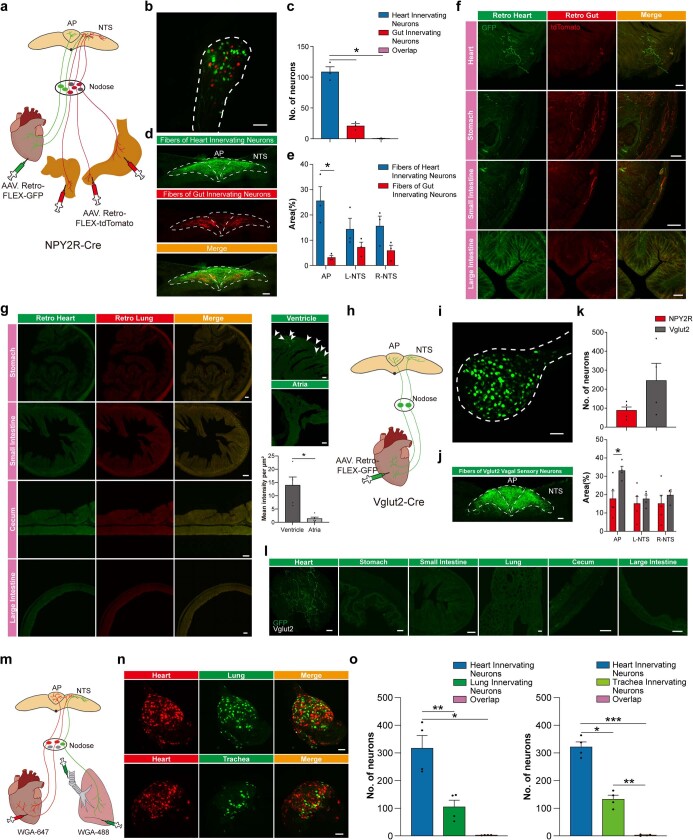
Extended Data Fig. 2. One-to-one map of vagal sensory neurons (VSNs).
a, Schematic of dual retrograde tracing of NPY2R VSNs from heart ventricles and gut (stomach, small and large intestine). b, Retrogradely labeled NPY2R VSNs from the heart ventricles (green), or gut (red, n = 3), in the nodose. c, Quantification of overlap. Nearly all of the heart and gut innervating neurons are non-overlapping, indicating that specific NPY2R VSNs project to the heart and gut separately (n = 3, heart/overlap p = 0.0112, heart/gut p = 0.0115). d, Spatial projection pattern of heart ventricles and gut innervating NPY2R VSNs in the AP and NTS (n = 3). e, Fiber density from heart ventricles or gut innervating NPY2R VSNs in the AP and NTS. The majority of fibers in the AP originate from heart ventricle innervating NPY2R VSNs (n = 3, p = 0.0102). f, NPY2R VSN terminals in retro-labeled heart ventricles and gut. Retro-labeled heart ventricle or gut fibers were only observed within heart ventricle or gut respectively (n = 3). g, NPY2R VSN terminals were not observed in stomach, small intestine, cecum, and large intestine after heart ventricle/lung dual retrograde tracing (n = 5). NPY2R VSN terminals were predominantly observed in the ventricle after atria/ventricle dual retrograde tracing (right, n = 5, p = 0.0145). h, Schematic of retrograde labeling (AAVrg-FLEX-GFP) of Vglut2 VSNs from the heart ventricles. Vglut2 is expressed by all VSNs. i, Retrogradely GFP labeled Vglut2 VSNs from the heart ventricles in the nodose ganglia (n = 4). j, Fiber distribution of retrogradely labeled GFP expressing Vglut2 VSNs from the heart ventricles in the AP and NTS (n = 4). k, Comparison of retrogradely labeled VSNs from the heart ventricles in Vglut2-Cre and NPY2R-Cre animals (top). Quantification of fibers from the heart ventricles innervating Vglut2 or NPY2R VSNs in the AP and NTS (bottom, n = 4 for Vglut2, n = 5 for NPY2R, p = 0.0225). l, Retrogradely labeled Vglut2 VSN terminals from the heart ventricles. Terminals were not observed in the lung, stomach, small and large intestine, or cecum (n = 4). m, Schematic of retrograde tracing of VSNs with wheat germ agglutinin (WGA) from heart ventricles (WGA 647), lung or trachea (WGA 488, n = 4). n, Heart ventricle (red), lung or trachea (green) innervating VSNs in the nodose ganglia after retrograde labeling (n = 4). o, Quantification of overlap after dual retro labeling. Similar to retrograde AAV tracing in NPY2R-Cre mice (Fig. 1e–g), most of the WGA488 and WGA647 labeled VSNs did not overlap (n = 4, heart/lung p = 0.0056, heart/overlap p = 0.0134; heart/trachea p = 0.0110, heart/overlap p = 0.0008, trachea/overlap p = 0.0068). p < 0.05* by one-way repeated measures ANOVA with Geisser-Greenhouse correction and Tukey multiple comparisons, two-way repeated measures ANOVA with Šidák multiple comparisons or two-way ANOVA with Šidák multiple comparisons. All error bars show mean ± s.e.m. Scale bars, 100 μm.