Abstract
Coronarin (COR), an analog of jasmonic acid, has been shown to enhance the tolerance of plants to drought. However, the effects of COR on the interactions among microorganisms associated with plant roots and their implications for enhancing the drought tolerance of plants remain unclear. Here, we studied the effects of applying COR on the microorganisms associated with plant roots and the rhizosphere metabolome. Treatment with COR affected the fungal community of the rhizosphere by inducing changes in the rhizosphere metabolome, which enhanced the drought tolerance of plants. However, treatment with COR had no significant effect on root microorganisms or rhizosphere bacteria. Specifically, the application of COR resulted in a significant reduction in the relative abundance of metabolites, such as mucic acid, 1,4-cyclohexanedione, 4-acetylbutyric acid, Ribonic acid, palmitic acid, and stearic acid, in maize roots under drought conditions; COR application also led to increases in the abundance of drought-resistant fungal microorganisms, including Rhizopus, and the assembly of a highly drought-resistant rhizosphere fungal network, which enhanced the drought tolerance of plants. Overall, the results of our study indicate that COR application positively regulates interactions between plants and microbes and increases the drought tolerance of plants.
Keywords: Coronarin, Drought stress, Rhizosphere microbiome, Rhizosphere metabolites
Graphical Abstract
1. Introduction
The continual increase in anthropogenic activities is altering the composition of the global atmosphere, and this has resulted in climate change [1], which poses a major threat to food security and agricultural production. Plants are exposed to unfavorable conditions at a greater frequency under global climate change, and this is expected to have a major effect on the production of crops [2]. Abnormal climate fluctuations can lead to irregular recipitation patterns, including drought and floods, and fluctuations in climate are expected to become more pronounced in future decades. Drought greatly limits agricultural production and affects crop quality and yield [1]. Drought stress can have major deleterious effects on the physiology of plants throughout their reproductive lives, including negative, irreversible effects. Water scarcity affects the uptake and transport of mineral nutrients in plants and numerous key metabolic pathways, leading to an increase in free radical levels and severe oxidative stress. In addition, prolonged and sustained drought disrupts the opening and closing of the stomata and gas exchange, results in the degradation of photosynthetic organs, and leads to alterations in the structure of the root system of plants [3].
Abscisic acid (ABA) is a common plant hormone that plays a key role plant drought tolerance [4]. ABA regulates the plant water balance by controlling the size of the stomatal aperture; it also induces other stress-related responses, including the accumulation of compatible solutes and antioxidant enzymes, as well as the release of proline [4]. It also plays a role in regulating root growth, carbohydrate storage, and the mobilization of stored proteins and amino acids [5]. Other hormones have been shown to enhance the drought tolerance of plants. Previous studies have shown that the drought tolerance of Arabidopsis is greater when the content of endogenous auxins is high, and this is associated with the positive effects of endogenous auxins on the structure of the root system [6]. Certain levels of cytokinins and ABA in plants under drought conditions can inhibit the growth of the lateral roots and promote the growth of the primary roots to facilitate water acquisition [7]. Jasmonic acid (JA) also plays a key role in the response to drought stress. The results of previous studies suggest that the application of exogenous JA can enhance the antioxidant activity of wheat and melon plants under drought conditions and reduce the deleterious effects of drought stress on plants. Coronatine (COR) is a non-host-specific phytotoxin produced by Pseudomonas syringae that is markedly similar to JA in its molecular structure and function [8]. Several studies have shown that foliar spraying of COR can enhance the drought tolerance of various plants, such as rice [9], wheat [10], tobacco [11], and maize [12]. COR is known to enhance plant drought tolerance by inducing the closure of the stomata and regulating reactive oxygen species (ROS) homeostasis, which decreases water loss and the activities of antioxidant enzymes [13], [14]. In addition, COR induces the expression of genes related to the biosynthesis of JA, including lipoxygenase, propylene oxide cyclase, and 12-oxygen-phytodienate reductase [13]. These signaling pathways play a role in systemic tolerance. An increasing number of studies have shown that COR can induce the production of anthocyanins in plants during drought stress [15], which suggests that COR plays a key role in regulating the plant microbiome. However, the role of interactions between COR-induced phytohormone signaling and plant-associated microbes in the drought tolerance of plants remains unclear.
The microorganisms surrounding plants can also play a role in increasing their drought tolerance [16]. For example, the abundance of mycorrhizal fungi increases under drought conditions, and this can enhance the water use efficiency of host plants via various routes [17]. Pretreatment of okra (Abelmoschus esculentus (L.) Moench) with Pseudomonas fluorescens solution has been shown to enhance drought tolerance [18]. The root-associated microbiome has also been shown to play a role in enhancing the drought tolerance of plants. The microbiomes of plants are often altered to enhance their tolerance to drought stress and growth conditions via “cry-for-help” strategies. Investigation of the complex feedback mechanisms between the host and microbes under drought stress is needed to clarify the role of the plant microbiome in drought tolerance. Furthermore, a study of the pathways involved in the recruitment and assembly of rhizosphere microbes under drought conditions can enhance our understanding of the recently proposed high yield–resource acquisition–stress tolerance (Y-A-S) theory [19] and help decrease crop losses induced by drought. COR, which is commonly used to mitigate the deleterious effects of drought on crops, has been shown to confer drought tolerance through studies of plant physiology. Thus, clarifying the role of plant–microbe interactions in enhancing drought tolerance is needed to promote the development of sustainable agricultural practices. Integrative studies of the mechanism by which COR confers drought tolerance and the plant microbiome might provide new insights that will aid the development of new strategies for enhancing the drought tolerance of plants and decreasing crop losses. Here, we collected microbes associated with maize roots following the foliar application of COR prior to and after drought. We also measured the plant rhizosphere metabolome under drought conditions following COR application. The specific aim of our study was to characterize the effects of the foliar application of COR on plant root-associated microbial communities.
2. Materials and Methods
2.1. Pot experiment
Soil was collected from the Baima Teaching and Research Base of Nanjing Agricultural University, Nanjing, Jiangsu Province, China. The “Zhengdan 958” maize variety was used in the pot experiment. Before commencing the experiment, preliminary assessments were conducted to ascertain the initial field capacity and moisture content of the soil, with measurements taken from a 100 g soil sample. Following surface sterilization with 10% sodium hypochlorite for 15 min, 150 seeds (75 seeds in each treatment) were allowed to germinate for 2 d in the dark at 25 ℃. One seed was planted per cell (6.8 cm × 7.5 cm, containing 100 g of soil) in the seeding trays (25 cells per tray), which were placed in a greenhouse with a 16 h light/8 h dark photoperiod (120 μmol photons m-2 s-1) with a 23 °C day/20 °C night temperature regime. Two mL of water was added to each cell every morning and evening. Once a week, 2 mL of 1/2 MS liquid medium was added to each cell instead of water. After one week, two treatments were established under normal moisture conditions: foliar spraying of COR (CORW) and foliar spraying of water (CKW). A total of 100 mL of 0.01 mol/L COR (purchased from Sangon Biotech (Shanghai) Co., Ltd.) was evenly applied to the foliage of 75 maize plants in the COR treatment group; an equal volume of water was sprayed on 75 plants in the control group (CK). COR was applied two times a week for two weeks. Six plants from each group under normal moisture conditions were harvested (CORW and CKW) for subsequent analyses. The remaining plants were then subjected to drought stress by maintaining the relative soil moisture content below 30% for 20 d. Specifically, prior to initiating the drought treatment phase, a one-week pre-drought period was imposed on the soil, during which the moisture content was closely monitored and measured. This preliminary phase aimed to ensure that the soil's moisture content remained below 30%, effectively replicating the drought conditions typically encountered by plants in their natural growth environment. Throughout the drought treatment period, meticulous control over the quantity of irrigation was maintained, with the goal of consistently keeping it within the narrow range of 25–26% of the calculated moisture requirement. Six plants from each replication under drought conditions (CORD and CKD) were randomly selected and used for measurements of biomass, chlorophyll content, proline content, and hydrogen peroxide (H2O2) levels and the collection of rhizosphere soil.
A portable chlorophyll meter SPAD-502 (Minolta, Japan) was used to measure the relative chlorophyll content of the leaves.
The procedure described by Monreal et al. [20] with some modifications was used to determine the content of proline. Fresh samples (0.5 g) were ground with 3% sulphosalicylic acid (5 mL) and then centrifuged. The supernatant (2 mL) was mixed with the same volume of acetic acid and acid ninhydrin (3 mL); the mixture was then incubated in an oven at 100 °C for 40 min, and the reaction was terminated in an ice bath. Toluene extraction (5 mL) of the reaction mixture was performed, and the absorbance was read at 520 nm using toluene as a blank control. The concentration of proline was calculated from a standard curve on a fresh weight basis.
Leaf samples (0.5 g) were homogenized at 4 °C in 8 mL of 50 mM ice-cold phosphate buffer (pH 7.0) containing 1% (w/v) polyvinylpyrrolidone. The homogenates were centrifuged at 15,000 g for 20 min at 4 °C, and the supernatants were collected and stored at − 80 °C until subsequent analyses. The H2O2 content was measured following the method described by Xie et al. [14].
2.2. Rhizosphere soil and root genome DNA extraction and sequencing
A Fast SoilPure DNA Isolation Kit (BOLAZ Biotech) was used to extract DNA from total rhizosphere soil and root samples per the manufacturer’s protocol. A NanoDrop 1000 spectrophotometer (Thermo Scientific, USA) and 1.2% agarose gel electrophoresis were used to measure the quality and quantity of the extracted DNA.
The Illumina MiSeq platform at Guangdong Magigene Biotechnology Co., Ltd., China, was used to sequence the ITS1–2 and V4 regions of the 16 S rRNA gene libraries. Primer pairs for bacteria (515 F: GTGYCAGCMGCCGCGGTAA; 806 R: GGACTACNVGGGTWTCTAAT) and fungi (ITS1F: CTTGGTCATTTAGAGGAAGTAA; ITS2R: GCTGCGTTCTTCATCGATGC) were used for amplification. Separate barcoded PCR amplicon libraries were built for bacterial and fungal community analysis. Procedures for the preparation of the bacterial and fungal amplicon libraries and high-throughput sequencing were based on those described in Yuan et al. [21].
The sequencing data were analyzed using usearch (V. 10.1) and vsearch (V. 0.6.3). First, the “vsearch --fastq_mergepairs” script was used to merge paired-end sequences; the “vsearch --fastx_filter” script was used to cut primers; the “vsearch --derep_fulllength” script was used to find unique read sequences; and the “usearch -unoise3” was used to generated ASVs; the “vsearch --usearch_global” script was used to create an ASV table. The “usearch -otutab” script was used to create an OTU table. The “vsearch --syntax” script, silva taxonomy database, and unite taxonomy database were used to annotate the bacterial and fungal OTUs; and the “vsearch --sintax” script and RDP taxonomic database were together used for annotation of representative sequences. A normalized number of sequences was randomly extracted from each sample in order to calculate alpha diversity indices that were estimated with the vegan R package[22].
For taxonomic annotations, representative sequences in the gene catalog were searched against the non-redundant protein database of NCBI with an e-value cutoff of 1e−5 using DIAMOND and the lowest common ancestor method was applied to estimate the assignment of genes to specific taxa.
Raw sequence data obtained in this study have been deposited in the Genome Sequence Archive in the BIG Data Center, Chinese Academy of Sciences under the accession (CRA011435).
2.3. Extraction of the rhizosphere soil metabolites
The metabolites from the rhizosphere soil were extracted and analyzed using a protocol adapted from the methodology outlined in studies [23], [24]. Briefly, each soil sample was split into two equal parts (0.2 g each) in two 2 mL Eppendorf (EP) tubes, and 24 μL of adonitol (1 mg.mL−1) was added to each tube as an internal standard. One part of the rhizosphere soil was homogenized in 0.5 mL methanol solution (Vmethanol: VH2O = 3:1) using a ball mill at 45 Hz for 4 min and then ultrasonically treated 5 times for a period of 5 min each.
The soil was then centrifuged at 10,000 g at 4 °C for 15 min, and the supernatant (∼0.4 mL) was transferred to a fresh EP tube. A second extraction was performed with 0.5 mL ethyl acetate using the same method as above, and the resultant extracts were combined (∼0.8 mL in total). Then, 0.5 mL of ethyl acetate and 0.5 mL of methanol solution were applied to each soil sample at the end of the extraction procedure. A second portion of soil sample was extracted by another 0.5 mL ethyl acetate followed by 0.5 mL methanol solution (Vmethanol: VH2O = 3:1) using the same method, resulting in an additional extract (∼0.8 mL). After four extractions, each sample yielded 1.6 mL solution. The extractions were analyzed by BIOTREE technology Co. Ltd. (Shanghai, China) using a gas chromatograph (Agilent 7890) coupled with time-of-flight mass spectrometry (GC-TOF-MS) as per the manufacturer’s instructions. Raw data processing and analyses were performed as previously reported. Briefly, Chroma TOF 4.3X software of the LECO Corporation and the LECO-Fiehn Rtx5 database were used for raw peaks exactions, data baseline filtering, calibration of the baseline, peak alignment, deconvolution analysis, peak identification and integration of the peak area. Metabolites were identified using both mass spectrum and retention index matches.
2.4. Statistical analysis
Statistical analyses were conducted using R 4.1 software [25]. Wilcoxon signed-rank tests were used to determine the significance of differences in plant physiological parameters among treatments. The threshold for statistical significance was adjusted p-value < 0.05. The Benjamini-Hochberg FDR procedure was employed to correct p-values for multiple comparisons. The R package “ggplot2” was used to make figures to visualize the results [26].
The R package “vegan” was used to calculate alpha diversity indices, which were determined from normalizedsequence counts that were randomly extracted from each sample [27]. Wilcoxon non-parametric tests were conducted to evaluate the significance of differences in the Shannon diversity, Pielou evenness, and Chao1 indicex among treatments using the “EasyStat” package. Prior to calculating beta diversity, the OTU profiles were standardized using relative abundance data. The “vegan” R package was used to build Bray–Curtis similarity matrices. Permutational multivariate analysis of variance (Adonis, transformed data by Bray-Curtis, permutations = 999) was used to determine whether beta diversity differed among treatments. Non-metric multidimensional scaling (NMDS) plots were generated using Bray–Curtis similarity matrices, which were created with the R package “ggplot2” [28]. The R package “edgeR” was used to identify differentially abundant microbes between treatments [29]. Network analysis was conducted using the R package “ggClusterNet” [30]. The network modules were computed using the “cluster_fast_greedy()” within the R package“igraph”.[31]. Based on the topological characteristics of nodes, node attributes can be categorized into four types, including Module hubs (nodes with high within-module connectivity within a module, Zi > 2.5 and Pi < 0.62), Connectors (nodes with high among-module connectivity bridging two modules, Zi < 2.5 and Pi > 0.62), Network hubs (nodes with high connectivity across the entire network, Zi > 2.5 and Pi > 0.62), and Peripherals (nodes with low connectivity both within and among modules, Zi < 2.5 and Pi < 0.62).
3. Results
3.1. Effect of COR application on plant growth
Foliar sprays of COR had no significant effect on the dry and fresh weight of the belowground biomass. The fresh weight and dry weight of the aboveground biomass of maize were approximately 50% and 54% higher, respectively, in the CORD treatment (COR application under drought conditions) than in the CKD treatment (control group under drought conditions) (Fig. 1A). No significant increase in the SPAD values (the relative chlorophyll content) of maize leaves was observed in the CORW treatment (COR application under non-drought conditions). The chlorophyll content of maize was 43% higher in the CORD treatment than in the CKD treatment, and this difference was significant (Fig. 1B). The proline content was similar in the COR and CK treatments when the humidity was appropriate. The proline content was 47% higher in the CORD treatment than in the CKD treatment (Fig. 1C). No differences in H2O2 concentrations were observed between the CORW and CKW treatments (control group under non-drought conditions)·H2O2 concentrations were 24% lower in the CORD treatment than in the CKD treatment (Fig. 1D).
Fig. 1.
: Effect of COR on maize growth and physiologicalperformance. A: Impact of COR application on maize biomass (both above and below-ground) under drought conditions. B: Effects of COR application on maize SPAD values before and after drought stress. Effect of COR on (C) proline content (D) H2O2 content. CKW, the control treatment (water spray) on normal water conditions. CKD, the control treatment on drought conditions. CORW, the COR application treatment on normal conditions. CORD, the COR application treatment on drought conditions. Each value represents the mean ± SD (n = 6). Bars showing the same letter are not significantly different at P ≤ 0.05 as determined by the LSD test.
3.2. COR application affected the root microbial communities of maize
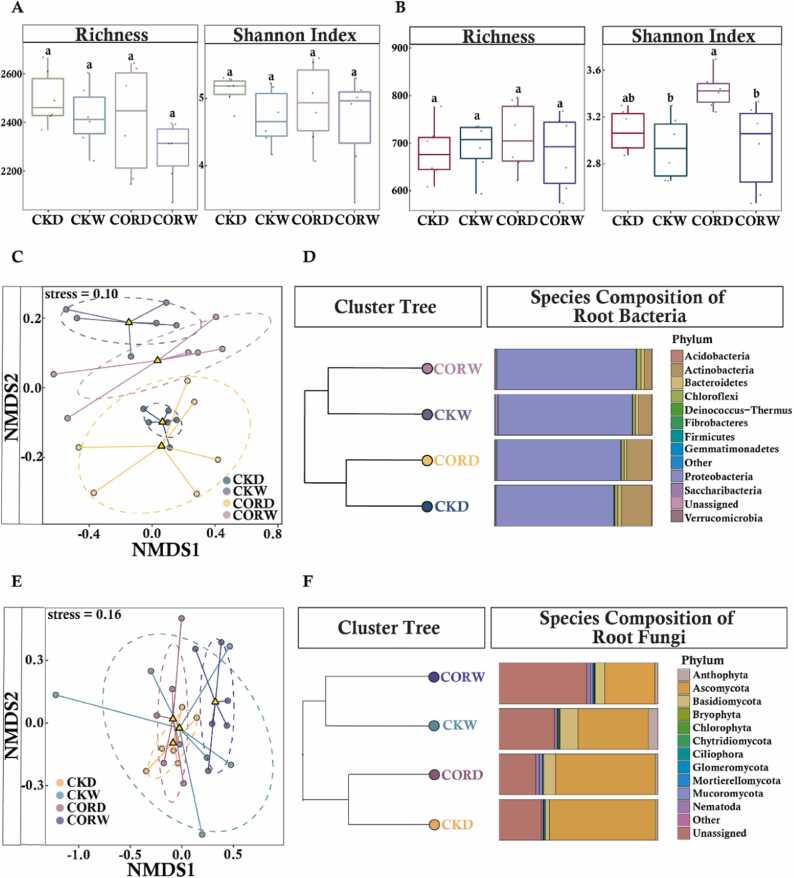
No significant differences in the richness and diversity of the root bacterial communities were observed among any of the treatments (Fig. 2A). There were no significant differences in fungal richness among treatments. Although the Shannon index and evenness of the fungal community were higher in the COR treatments than in the other treatments, no significant differences in the alpha diversity of fungi in maize roots were observed among treatments (Fig. 2B). Non-metric multidimensional scaling (NMDS) analysis was conducted to compare the similarity in the composition of the microbial community among treatments. Drought had a major effect on the clustering patterns of the bacterial community of the maize roots. However, regardless of the water conditions, bacterial communities in the COR treatment and control were similar (Fig. 2C). No significant differences were observed in root fungal communities among treatments (Fig. 2D). Distances in the clustering tree indicate group heterogeneity, with lower distances indicating greater similarity. Significant variation was observed in the species composition of the bacterial and fungal communities in maize roots under drought and normal moisture conditions. Root fungal and bacterial communities primarily cluster based on moisture conditions. This shows that COR application had a weaker effect on the composition of species than moisture conditions (Fig. 2E). The average relative abundances of taxa in the four groups were determined; Proteobacteria was the most abundant bacterial phylum across all treatment groups. The relative abundance of Actinobacteria in the roots of maize was higher under drought conditions than under normal moisture conditions, and COR application mitigated this increase. The relative abundance of another phylum of maize root bacteria, Saccharibacteria, was significantly lower under drought conditions than under normal moisture conditions, and COR application mitigated this decrease. The relative abundance of Ascomycota was significantly higher under drought conditions than under normal moisture conditions. The relative abundance of Basidiomycota in maize roots was significantly lower in the CKD treatment than in the CORD treatment (Fig. 2F). The relative abundance of Basidiomycota in maize roots was significantly higher in the CORW treatment than in the CKW treatment, and no differences in the relative abundance of Basidiomycota were observed under drought and normal moisture conditions. These findings suggest that the foliar application of COR does not have a strong effect on maize root microbes.
Fig. 2.
: The diversity and composition of maize root microbial communities between COR application and control treatment under different moisture conditions. Bacterial (A) and fungal (B) communities were assessed using various alpha diversity indices for all clustered OTUs. The horizontal bars within the boxes represent the medians. The tops and bottoms of boxes represent the 75th and 25th quartiles, respectively. All outliers were plotted as individual points. Non-metric multidimensional scaling (NMDS), based on Bray-Curtis dissimilarities, was conducted on the taxonomic profile (OTU level) of bacterial (C) or fungal (D) communities under different treatment conditions. Relative abundance (%) of members within the bacterial (E) and fungal (F) phyla was also determined. The clustering tree hierarchically groups the samples using K-means clustering method and represents the data distances between groups of samples.
3.3. COR application affected the rhizosphere microbial communities of maize
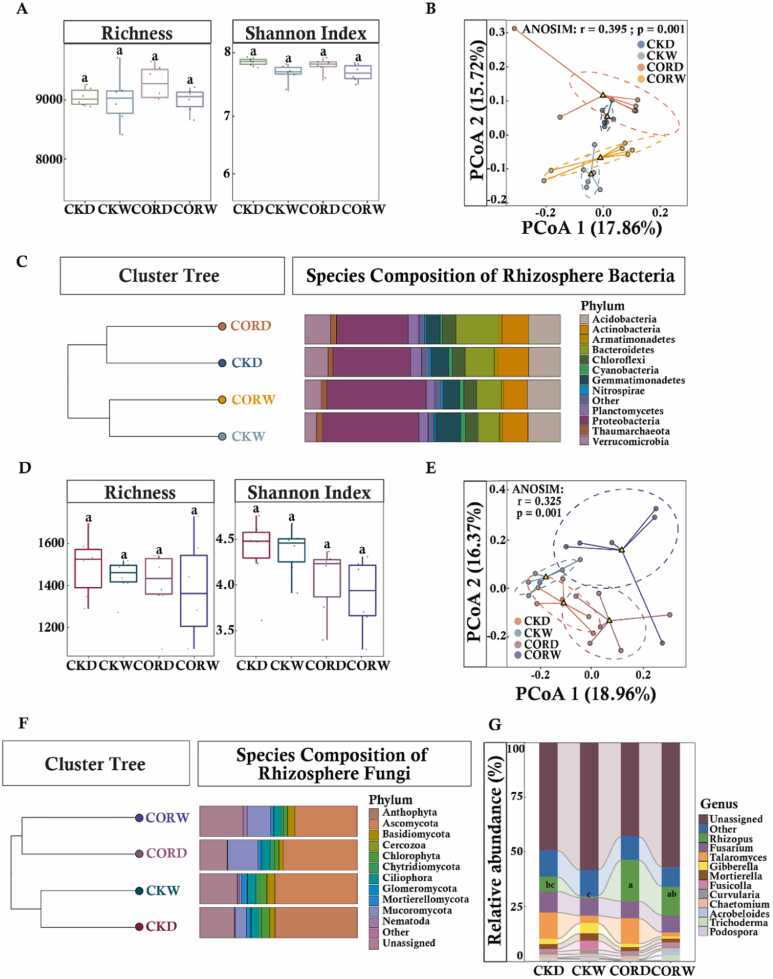
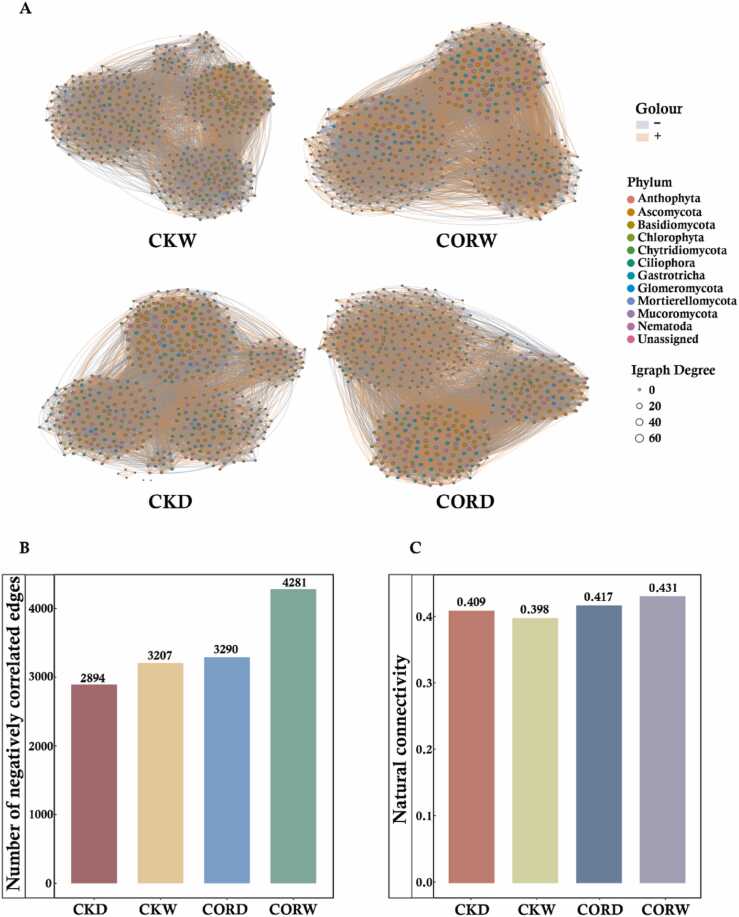
No significant variation in the Alpha diversity of rhizosphere bacteria, including richness, Shannon index, and evenness, was observed among treatments. Principal coordinate analysis (PCoA) based on Bray–Curtis distance matrices revealed that moisture conditions affected the rhizosphere bacterial community of maize, as indicated by the separation between the rhizosphere bacterial community under drought stress and normal moisture conditions (Fig. 3A). The clustering analysis revealed that rhizosphere bacterial communities were first clustered by moisture conditions (with low distances in the clustering tree) and then by COR application, which indicated that drought had a major effect on the maize rhizosphere bacterial community (Fig. 3B). The relative abundance of Bacteroidetes was significantly higher in the CORD treatment than in the CKD treatment. The relative abundance of Gemmatimonadetes in the maize rhizosphere bacterial community was lower in the COR treatments than in the CK treatments (Fig. 3C). No significant differences in fungal alpha diversity of the maize rhizosphere were observed among treatments (Fig. 3D). The clustering patterns of rhizosphere fungi differed in the CORW and CORD treatments (Fig. 3E), indicating that the structure of the rhizosphere fungal community differed under drought and normal moisture conditions. The rhizosphere fungal community was not completely separated in the CKW and CORW treatments but overlapped in the CKD and CORD treatments. This indicates that the structure of the rhizosphere fungal community was affected by both the application of COR and moisture conditions. A multifactorial analysis of rhizosphere fungi indicated that both COR application and moisture conditions had strong effects on the structure of the rhizosphere fungal community (Table 1). In the clustering tree of rhizosphere fungal species, the distance between CORW and CORD was smaller than that between CORW and the other groups, indicating that COR application had a greater effect on the clustering of rhizosphere fungal taxa than moisture conditions. The abundance of Mucoromycota was higher in the COR treatments than in the CK treatments. Specifically, the abundance of this phylum was 13.4% higher in CORW than in CKW and 12.1% higher in CORD than in CKD. These changes were mostly driven by variation in Rhizopus; the abundance of Rhizopus was higher in CORD than in the other treatments (Fig. 3F, G). The relative abundances of Ascomycota and Chlorophyta were lower in the COR treatments than in the CK treatments; the abundances of these phyla were 5.3% lower in the CORW treatment than in the CKW treatment and 12.6% lower in the CORD treatment than in the CKD treatment. The top 40 OTUs that made the largest contributions to the different groupings were identified as characteristic microorganisms via a machine learning random forest model of the rhizosphere fungal community (Table S1). We built a co-occurrence network based on the Spearman correlations between OTUs to analyze relationships among rhizosphere fungi (Fig. 4A). Topological comparisons of the networks revealed that the proportion of negative edges was higher in the COR treatment (Fig. 4B), which indicates that COR treatment can increase the stability of the rhizosphere fungal network. The number of natural connections remaining after randomly removing 50% of the nodes was determined to further evaluate community stability. After randomly removing 50% of the nodes, the number of natural connections was higher in the CORW treatment than in the CKW treatment; the number of natural connections was also higher in the CKD treatment than in the CKW treatment, suggesting that the application of COR might increase the stability of the rhizosphere fungal community (Fig. 4C). In addition, the nodes that are key to the network interactions, which were referred to as key microorganisms, were filtered using the ZiPi values of the network (Table S2).
Fig. 3.
: Effect of COR on the rhizosphere microbial community of maize under different moisture conditions. Alpha diversity of rhizosphere bacterial (A) among different treatments. Chao1 and Shannon indices were calculated using the normalized OTUs table. Horizontal bars within boxes represent the median. The tops and bottoms of boxes represent the 75th and 25th quartiles, respectively. Outliers were plotted as individual points. Bacterial (B) PCoA analysis of different treatments computed using species-level profiles and Bray-Curtis distances. Stack bar plot depicts the relative abundances (%) of major phyla present in the bacterial communities(C). D: Rhizosphere fungal alpha diversity. E: Fungal PCoA analysis of different treatments computed using species-level profiles and Bray-Curtis distances. F: Stack bar plot depicts the relative abundances (%) of major phyla present in the fungal communities. G: Stack bar plot depicts the relative abundances (%) of major genera present in the fungi communities. Bars showing the same letter are not significantly different at P ≤ 0.05 as determined by the LSD test.
Table 1.
Two-factor analysis of the effects of COR application and drought on rhizosphere fungal communities.
| Df | R2 | F | Pr | |
|---|---|---|---|---|
| COR Treatment | 1 | 0.1166 | 3.1611 | 0.001 * * |
| Water Conditions | 1 | 0.0999 | 2.7086 | 0.001 * * |
| Interaction Effects | 1 | 0.0458 | 1.2415 | 0.213 |
Fig. 4.
: Network Analysis of Maize Rhizosphere Fungal Communities. A: Co-occurrence networks OTUs in the maize rhizosphere fungi (> 1%). Edges represent significant Spearman correlations (ρ > |0.8|, P < 0.05). Light blue lines represent significant negative correlations, while light red lines represent significant positive correlations. Nodes represent OTUs, and edges represent significant interactions among nodes. B: The proportion of negative edges in the maize rhizosphere fungal network. C: Natural connectivity of the rhizosphere fungal network of maize for each treatment following random removal of 50% of the nodes.
3.4. Effect of COR application on rhizosphere metabolomics
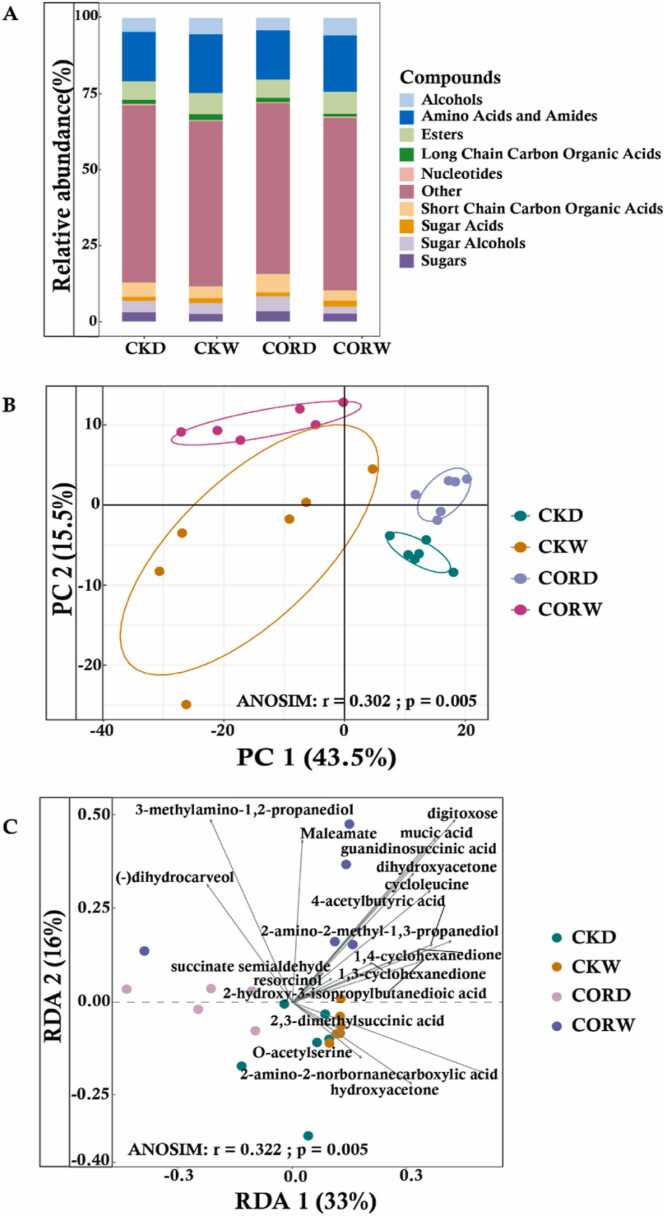
To explore the mechanism underlying the effects of the foliar application of COR on the tolerance of plants to drought stress, gas chromatography–mass spectrometry (GC–MS) was used to characterize rhizosphere metabolic profiles. Analysis of the rhizosphere metabolome by GC–MS yielded 750 chromatographic peaks. A total of 112 compounds were identified, including 12 alcohols, 12 amino acids and amides, 9 esters, 4 long-chain carbon organic acids, 1 nucleotide, 12 short-chain carbon organic acids, 3 sugar acids, 2 sugar alcohols, 9 sugars, and 48 other components. We identified the rhizosphere metabolites of each group, and our findings revealed significant decreases in the percentage of alcohols, amino acids and amides, sugar acids, sugar alcohols and esters by 0.76%, 3.07%, 0.41%, 0.2% and 0.19%, respectively, following COR application. A total of 15 metabolites with significant differences among groups were subjected to subsequent analyses. The relative abundance of sugars and short-chain carbon organic acids was 0.58% and 0.89% higher in rhizosphere soil under drought stress than under normal moisture conditions, respectively, and the relative abundance of short-chain carbon organic acids and sugar alcohols was higher in the CORD treatment than in the CKD treatment (Fig. 5A). Multivariate PCA was conducted on all metabolites to reduce the dimensionality of the dataset.As many variables were correlated with each other, two determined major principal components (PC1 and PC2)could explain 59.0% of the total variance, i.e., 43.5% and 15.5% for PC1 and PC2, respectively (Fig. 5B). PCA plots using K-mean clustering revealed two major groups of metabolite profiles corresponding to drought and non-drought conditions. All groups on the PCA plot were split into drought and non-drought clusters, which revealed significant differences in the identified metabolites in each treatment (Fig. 5B). A redundancy analysis was conducted to determine the extent to which COR application and drought are associated with changes in the plant rhizosphere metabolome. The results revealed that four metabolites, O-acetyl serine, 2,3-dimethyl succinic acid, 2-amino-2-norbornanecarboxylic acid, and hydroxyacetone, were the major contributors to differences in the maize rhizosphere metabolome between the control and CORW treatments. The rhizosphere metabolomes of plants separated in drought and non-drought environments; 3-methylamino-1,2-propanediol and dihydrocarveol were correlated with the CORD treatment, and maleamate, succinate semialdehyde, resorcinol, cyclohexanedione, guanidino succinic acid, mucic acid, digitoxose, dihydroxyacetone, cycloleucine, 2-amino-2-methyl-1,3-propanediol, and 2-hydroxy-3-isopropylbutanedioic acid were correlated with the CORW treatment (Fig. 5C). These findings indicate that both COR treatment and drought conditions affected plant rhizosphere metabolomes. A machine learning random forest model was used to determine the 40 metabolites that made the largest contributions to differences among groups, and these were designated as key metabolites in subsequent analyses (Table S3).
Fig. 5.
: Effect of COR application on the rhizosphere metabolome. A: Profiling of the rhizosphere metabolome. Metabolites are categorized based on their chemical properties and represented using stacked column charts. The lowerase letters inserted indicate significant differences in their relative abundances (P ≤ 0.05). B: Principal Component Analyses (PCA) of rhizosphere soil metabolome by GC–MS (adonis, r2 = 0.84, P < 0.01). C: Redundancy analysis (RDA) of the abundant fungal phyla and rhizosphere metabolome under different treatments conditions.
3.5. Interaction of metabolites with the rhizosphere fungal community
The aforementioned results indicate that the application of COR had a weak effect on root microbes (both bacteria and fungi) and rhizosphere bacterial communities under drought and normal moisture conditions. Conversely, COR application had a significant effect on the fungal community of the maize rhizosphere. To clarify the effects of the foliar application of COR on the metabolome and rhizosphere fungal communities under drought conditions, we first explored the relationship between highly abundant rhizosphere fungal genera and differential metabolites (Fig. 6A). The abundances of Acrobeloides and Trichoderma were considerably higher in the CORW treatment than in the CKW treatment. These two taxa consistently responded to key metabolites in the rhizosphere, and both were positively correlated with metabolites such as mucic acid, 1,4-cyclohexanedione, 4-acetylbutyric acid, Ribonic acid, palmitic acid and stearic acid. Fusicolla was significantly more abundant in the CKW treatment than in the CORW treatment; it was also positively correlated with the above key metabolites (Fig. 6A). Rhizopus was more abundant in the CORD treatment than in the CKD treatment; however, Rhizopus was negatively correlated with the aforementioned compounds (Fig. 6A). The abundances of Talaromyces and Rhizopus were significantly higher in the CKD and CORD treatments than in the CKW and CORW treatments; changes in these taxa in response to key metabolites were essentially the same (Fig. 6A). Key metabolites and rhizosphere fungal co-occurrence networks at the OTU level revealed the effects of key metabolites on relationships in the fungal network. Most OTUs clustered around key metabolites belonging to the ZiPi of the CORD and CORW treatments (Fig. 6B).
Fig. 6.
: A: Correlation between rhizosphere key metabolites and high abundance of fungal genera. Red colour means a positive correlation and blue colour means a negative correlation. B: Rhizosphere key metabolites and fungal co-occurrence networks. Red nodes represent fungal OTUs, and green nodes denote key metabolites. Light blue lines represent significant positive correlations and light red lines represent significant negative correlations. C: Selection methods for key rhizosphere fungi in the CORD and CKD groups. D: Conceptual diagram with the partial least-squares path model illustrating the promotion of plant growth under drought conditions. The PLS-PM shows both direct and indirect ways which rhizosphere metabolites influence plant rhizosphere microbial communities and plant physiological indicators. The red lines indicate negative links, and the blue lines represent positive correlations. Arrow widths correspond to the relative effect size of each variable. Numbers associated with each arrow indicate path coefficients (P = 0.05). R2 indicates variance indices explained by the model. * *, 0.001 < P < 0.01.
3.6. COR application affected the rhizosphere microecology of maize
We also conducted partial least squares path modeling (PLS-PM) analysis to clarify the potential linkages among key rhizosphere fungi, rhizosphere metabolites profile, and plant growth properties. The physiological indicators of maize examined were biomass, chlorophyll content, proline content, and H2O2 content, collectively prociding a comprehensive measure of plant drought. We obtained ZiPi values, which indicated the most important OTUs in each group, by measuring the properties of the networks. We then compared these OTUs with the Top 40 OTUs obtained by the machine learning random forest model of the rhizosphere fungal community in the previous phase, and overlapping OTUs were identified as the key microorganisms in each group (Fig. 6C). The application of COR had a significant effect on rhizosphere metabolites profile, the key fungal groups, and the drought tolerance of plants. The positive correlation between COR treatment and plant drought tolerance indicated that the application of COR enhanced the drought tolerance of plants. COR application affected the rhizosphere metabolome profile and key microorganisms in the rhizosphere fungal community under drought conditions. Moreover, rhizosphere metabolites profile were significantly negatively correlated with the key microorganisms of CKD and positively correlated with the key microorganisms of CORD and plant drought tolerance. These results indicate that rhizosphere metabolites profile affect the entire rhizosphere microbial community through their effects on key fungal microorganisms, ultimately increasing the tolerance of plants to drought (Fig. 6D).
4. Discussion
Coronatine is a structural and functional imitator of jasmonates that alters the physiological and biochemical reactions of plants under stress conditions [32]. COR application significantly increased the aboveground biomass of maize plants and mediated the maintenance of high SPAD values under drought conditions. Drought stress decreased the chlorophyll content, which might inhibit the accumulation of dry matter in maize plants. This is consistent with the results of previous studies suggesting that COR application increased the dry matter content, the content of chlorophyll, and the photosynthetic rate of leaves under drought stress [8]. The application of COR in the aforementioned study [13] has been shown to result in a decrease in the content of H2O2, which might lead to increases in antioxidant activity that facilitate the elimination of ROS and alter the proportions of membrane fatty acids to decrease vulnerability to ROS attack. Proline accumulation is an indicator of drought stress tolerance in plants. The accumulation of proline is thought to reduce cell osmotic potential and induce the absorption of water by plants to regulate the osmotic balance [20]. Our results indicate that the content of proline was significantly higher in maize plants treated with COR under drought stress than in untreated maize plants. In sum, our results indicate that COR application can mitigate the adverse effects of drought stress on maize growth.
Our results indicate that under drought conditions, the application of COR does not significantly affect the maize roots microbiota. Drought significantly increased the relative abundance of rhizosphere bacteria, such as Bacteroidetes and Verrucomicrobia in maize, which is consistent with the results of Sheik [33] and Liu [34]. Verrucomicrobia is associated with polysaccharide metabolism [35], and the abundance of Verrucomicrobia is dependent on the dead leaves of plants and root secretions. Bacteroidetes plays a key role in the ability of plants to tolerate extreme conditions. Rhizosphere microbial analysis revealed that maize rhizosphere fungi are more strongly affected by COR treatment than drought conditions. The abundance of Rhizopus genus was higher in the CORD treatment than in the CKD treatment, and a previous study has indicated that Rhizopus can enhance the drought tolerance of plants [36]. Soil fungal communities are more stable under drought conditions than soil bacterial communities [37], indicating that soil fungal communities are more drought resistant than soil bacterial communities. Moreover, recent studies have shed light on the intricate dynamics of soil microbial communities in response to drought stress.The results of Hasibeder et al. [38] have shown that drought might alter the allocation of carbon to the subsurface, and the results of Fuchslueger et al. [39] have shown that the allocation of carbon to bacteria, but not fungi, is reduced in mountain grasslands, which might explain the potential positive response of plant rhizosphere fungi under drought stress. Our results indicated that COR application can enhance the stability of the rhizosphere fungal network. High stability of the structure of the microbial network ensures that the entire microbial system remains functional after disturbance. This might be one of the mechanisms by which COR enhances the tolerance of plants to drought stress.
The abundance of rhizosphere soil microbes is thought to increase following root exudation, and certain bacteria thrive in the rhizosphere based on changes in rhizosphere metabolites [40]. Our findings indicate that, following drought conditions, the application of COR treatment resulted in a significant increase in the relative content of sugars and short-chain organic acids within the rhizosphere metabolome compared to the control. Although we previously found that sugars and short-chain organic acids have inhibitory effects on the growth and recruitment of beneficial bacteria, some of these substances were beneficial to the growth of specific fungi. Cellobiose [41], mucic acid [42], 2-deoxy-D-glucose [43], succinic acid [44], glycolic acid [45] and fructose [46] were important metabolites of the rhizosphere in our study. According to our results, the COR treatment exhibited the effect on the rhizosphere metabolomes variationThe tolerance of plants to stress is enhanced via increases in the root exudation of 2,3-dimethyl succinic acid [47], and Huang et al. [47] demonstrated the correlation of 2,3-dimethyl succinic acid exudation with rhizosphere fungal communities. These key metabolites are positively correlated with some of the beneficial fungi with increased abundances in the COR treatment. Our results indicate that rhizosphere metabolites recruit key rhizosphere fungi and that this change is caused by COR application. Many studies have shown that drought-induced changes in root exudation patterns affect rhizosphere physiological processes and can potentially affect ecosystem responses [48]. Rhizosphere metabolites can select for beneficial soil microbial communities. Plants need to balance the needs for production and defense under drought stress, and this has important implications for the biodiversity and function of rhizosphere ecosystems. The plant rhizosphere metabolome plays a key role in the rhizosphere ecosystem under drought, and COR application mediates the recruitment of fungal microorganisms with the ability to stabilize communities under drought by altering the plant rhizosphere metabolome to compensate for the weakened defenses of plants under drought conditions.
Due to the influence of plant age on various physiological and metabolic processes, our study acknowledges the potential variability in differences pertaining to plant-associated microorganisms and metabolites under both drought and non-drought conditions. Nonetheless, it is cruxial to note that significant and noteworthy disparities persist in both rhizospheric fungal community structure and metabolite profiles between two groups of plants (applicated COR or not) on the same age exposed to drought conditions.The effects of COR application on the plant, rhizosphere microbiome, and rhizosphere metabolomics remain unclear because of various confounding factors under drought stress. We attempted to identify the processes associated with the ability of COR application to enhance the tolerance of the plant rhizosphere to drought stress via PLS-PM analysis. PLS-PM analysis revealed that COR application significantly affected rhizosphere metabolites, which affects the entire rhizosphere microbial community, especially key fungal microorganisms, and ultimately enhances the tolerance of plants to drought. The results of this study are the first to show that the foliar application of COR improves the tolerance of the plant rhizosphere to drought by altering the plant rhizosphere metabolome, which affects key rhizosphere fungi.
5. Conclusion
Our study has revealed the significant impact of COR application on the maize rhizosphere fungal community, particularly under drought stress conditions. Notably, COR treatment resulted in a substantial increase in specific metabolites within the maize rhizosphere. Additionally, it induced changes in the structure and network of the fungal microbial community in the rhizosphere, including an increase in drought-tolerance fungi such as Rhizopus. These changes contributed to the formation of a more robust and resilient fungal network in the rhizosphere. In summary, our findings demonstrate that COR treatment profoundly influences the rhizosphere metabolome and the composition and functionality of the rhizosphere fungal community under drought conditions, ultimately enhancing the plant's drought tolerance. These insights into the intricate interplay between plant metabolomics and the rhizosphere microbiome hold great promise for the development of strategies aimed at bolstering plant resilience under adverse environmental conditions, potentially revolutionizing agricultural practices for sustainable crop production in the face of increasing environmental challenges.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was financially supported by Key Research and Development Projects of Henan Province (231111113000), Natural Science Foundation of China (42277297, 42322708), Natural Science Foundation of Jiangsu Province (BK20211577), the Fundamental Research Funds for the Central Universities (Grant No. KYT2023001, XUEKEN2023039, CGZH2023001) and the Innovative Research Team Development Plan of the Ministry of Education of China (Grant No. IRT_17R56). J. Y. was supported by Qing Lan Project of Jiangsu Province.
Author contributions
PD, XL, GN, NJ, TW, and JY: conducted all experiments; PD, XL and GN wrote the paper; JY and QS conceived the study, supervised the study; QC, JZ, JGZ and CX: provided critical comments on the study, and helped write the paper.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.10.043.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Vicente-Serrano S.M., McVicar T.R., Miralles D.G., Yang Y., Tomas-Burguera M. Unraveling the influence of atmospheric evaporative demand on drought and its response to climate change. Clim Change. 2020;11 [Google Scholar]
- 2.Peng B., Guan K., Tang J., Ainsworth E.A., Asseng S., et al. Towards a multiscale crop modelling framework for climate change adaptation assessment. Nat Plants. 2020;6:338–348. doi: 10.1038/s41477-020-0625-3. [DOI] [PubMed] [Google Scholar]
- 3.Gupta A., Rico-Medina A., Caño-Delgado A.I.J.S. The physiology of plant responses to drought. Science. 2020;368:266–269. doi: 10.1126/science.aaz7614. [DOI] [PubMed] [Google Scholar]
- 4.Soma F., Takahashi F., Yamaguchi-Shinozaki K., Shinozaki K.J.P. Cellular phosphorylation signaling and gene expression in drought stress responses: ABA-dependent and ABA-independent regulatory systems. Plants. 2021;10:756. doi: 10.3390/plants10040756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu W., Jia L., Shi W., Liang J., Zhou F., et al. Abscisic acid accumulation modulates auxin transport in the root tip to enhance proton secretion for maintaining root growth under moderate water stress. New Phytol. 2013;197:139–150. doi: 10.1111/nph.12004. [DOI] [PubMed] [Google Scholar]
- 6.Salvi P., Manna M., Kaur H., Thakur T., Gandass N., et al. Phytohormone signaling and crosstalk in regulating drought stress response in plants. Plant Cell Rep. 2021;40:1305–1329. doi: 10.1007/s00299-021-02683-8. [DOI] [PubMed] [Google Scholar]
- 7.Calvo-Polanco M., Armada E., Zamarreño A.M., García-Mina J.M., Aroca R. Local root ABA/cytokinin status and aquaporins regulate poplar responses to mild drought stress independently of the ectomycorrhizal fungus Laccaria bicolor. J Exp Bot. 2019;70:6437–6446. doi: 10.1093/jxb/erz389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu H., Wu X., Li Z., Duan L., Zhang M. Physiological evaluation of drought stress tolerance and recovery in cauliflower (Brassica oleracea L.) seedlings treated with methyl jasmonate and coronatine. J Plant Growth Regul. 2012;31:113–123. [Google Scholar]
- 9.Ai L., Li Z., Xie Z., Tian X., Eneji A., et al. Coronatine alleviates polyethylene glycol‐induced water stress in two rice (Oryza sativa L.) cultivars. J Agron Crop Sci. 2008;194:360–368. [Google Scholar]
- 10.Li X., Shen X., Li J., Egrinya Eneji A., Li Z., et al. Coronatine alleviates water deficiency stress on winter wheat seedlings. Plant Sci Biomol. 2010;52:616–625. doi: 10.1111/j.1744-7909.2010.00958.x. [DOI] [PubMed] [Google Scholar]
- 11.Xu J., Zhou Y., Xu Z., Chen Z., Duan L. Physiological and transcriptome profiling analyses reveal important roles of coronatine in improving drought tolerance of tobacco. J Plant Growth Regul. 2020;39:1346–1358. [Google Scholar]
- 12.Yu H., Wang Y., Xing J., Zhang Y., Duan L., et al. Coronatine modulated the generation of reactive oxygen species for regulating the water loss rate in the detaching maize seedlings. Agriculture. 2021;11:685. [Google Scholar]
- 13.Hao L., Wang Y., Zhang J., Xie Y., Zhang M., et al. Coronatine enhances drought tolerance via improving antioxidative capacity to maintaining higher photosynthetic performance in soybean. Plant Sci. 2013;210:1–9. doi: 10.1016/j.plantsci.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Xie Z., Duan L., Tian X., Wang B., Eneji A.E., et al. Coronatine alleviates salinity stress in cotton by improving the antioxidative defense system and radical-scavenging activity. J Plant Physiol. 2008;165:375–384. doi: 10.1016/j.jplph.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Gao M., Zhang C., Lu H.J.S.R. Coronatine is more potent than jasmonates in regulating Arabidopsis circadian clock. Sci Rep. 2020;10 doi: 10.1038/s41598-020-69627-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ullah A., Nisar M., Ali H., Hazrat A., Hayat K., et al. Drought tolerance improvement in plants: an endophytic bacterial approach. Appl Microbiol Biotechnol. 2019;103:7385–7397. doi: 10.1007/s00253-019-10045-4. [DOI] [PubMed] [Google Scholar]
- 17.Badr Eldin R., G Saad M.M., KH Abdelhalim A.E. Using Fungal Endophytes for Increasing Water Productivity and Tolerance of Wheat Plants to Drought Stress. Alex Sci Exch J. 2022;43:711–718. [Google Scholar]
- 18.Pravisya P., Jayaram K., Yusuf A.J.P., Plants M.B.O. Biotic priming with Pseudomonas fluorescens induce drought stress tolerance in Abelmoschus esculentus (L.) Moench (Okra) Physiol Mol Biol Plants. 2019;25:101–112. doi: 10.1007/s12298-018-0621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malik A.A., Puissant J., Goodall T., Allison S.D., Griffiths R.I., et al. Soil microbial communities with greater investment in resource acquisition have lower growth yield. Soil Biol Biochem. 2019;132:36–39. [Google Scholar]
- 20.Monreal J., Jimenez E., Remesal E., Morillo-Velarde R., García-Mauriño S., et al. Proline content of sugar beet storage roots: Response to water deficit and nitrogen fertilization at field conditions. Environ Exp Bot. 2007;60:257–267. [Google Scholar]
- 21.Yuan J., Wen T., Zhang H., Zhao M., Penton C.R., et al. Predicting disease occurrence with high accuracy based on soil macroecological patterns of Fusarium wilt. ISME J. 2020;14:2936–2950. doi: 10.1038/s41396-020-0720-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oksanen J., Kindt R., Legendre P., O’Hara B., Stevens M.H.H., et al. The vegan package. Community Ecol Package. 2007;10:719. [Google Scholar]
- 23.Wen T., Zhao M., Liu T., Huang Q., Yuan J., et al. High abundance of Ralstonia solanacearum changed tomato rhizosphere microbiome and metabolome. BMC Plant Biol. 2020;20:1–11. doi: 10.1186/s12870-020-02365-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen T., Xie P., Liu H., Liu T., Zhao M., et al. Tapping the rhizosphere metabolites for the prebiotic control of soil-borne bacterial wilt disease. Nat Commun. 2023;14 doi: 10.1038/s41467-023-40184-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ihaka R., Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 26.Wickham H., Averick M., Bryan J., Chang W., McGowan L.D.A., et al. Welcome to the Tidyverse. J Open Source Softw. 2019;4:1686. [Google Scholar]
- 27.Worley B., Powers R. Multivariate analysis in metabolomics. Curr Metab. 2013;1:92–107. doi: 10.2174/2213235X11301010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wickham H.J.Wircs. ggplot2. Wiley interdisciplinary reviews: computational statistics. 2011; 3:180–185.
- 29.Robinson M.D., McCarthy D.J., Smyth G.K.J.B. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen T., Xie P., Yang S., Niu G., Liu X., et al. ggClusterNet: An R package for microbiome network analysis and modularity‐based multiple network layouts. iMeta. 2022;1 doi: 10.1002/imt2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.G. Csardi T.J.I. Nepusz complex systems igraph Softw Package Complex Netw Res 1695 2006 1 9.
- 32.Anjum S., Wang L., Farooq M., Khan I., Xue L. Methyl jasmonate-induced alteration in lipid peroxidation, antioxidative defence system and yield in soybean under drought. J Agron Crop Sci. 2011;197:296–301. [Google Scholar]
- 33.Sheik C.S., Beasley W.H., Elshahed M.S., Zhou X., Luo Y., et al. Effect of warming and drought on grassland microbial communities. ISME J. 2011;5:1692–1700. doi: 10.1038/ismej.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T.Y., Ye N., Wang X., Das D., Tan Y., et al. Drought stress and plant ecotype drive microbiome recruitment in switchgrass rhizosheath. J Integr Plant Biol. 2021;63:1753–1774. doi: 10.1111/jipb.13154. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Garcia M., Brazel D.M., Swan B.K., Arnosti C., Chain P.S., et al. Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PloS One. 2012;7 doi: 10.1371/journal.pone.0035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Badr Eldin R., G Saad M.M., KH Abdelhalim A.E. Using Fungal Endophytes for Increasing Water Productivity and Tolerance of Wheat Plants to Drought Stress. Alex Sci Exch J. 2022;43:711–718. [Google Scholar]
- 37.de Vries F.T., Griffiths R.I., Bailey M., Craig H., Girlanda M., et al. Soil bacterial networks are less stable under drought than fungal networks. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasibeder R., Fuchslueger L., Richter A., Bahn M. Summer drought alters carbon allocation to roots and root respiration in mountain grassland. N Phytol. 2015;205:1117–1127. doi: 10.1111/nph.13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuchslueger L., Bahn M., Fritz K., Hasibeder R., Richter A. Experimental drought reduces the transfer of recently fixed plant carbon to soil microbes and alters the bacterial community composition in a mountain meadow. N Phytol. 2014;201:916–927. doi: 10.1111/nph.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamakawa H., Hakata M. Atlas of rice grain filling-related metabolism under high temperature: joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant Cell Physiol. 2010;51:795–809. doi: 10.1093/pcp/pcq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Y., Yang M., Yin R., Wang L., Luo L., et al. Autotoxin Rg1 induces degradation of root cell walls and aggravates root rot by modifying the rhizospheric microbiome. Microbiol Spectr. 2021;9 doi: 10.1128/spectrum.01679-21. e01679-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shang H., Ma C., Li C., Zhao J., Elmer W., et al. Copper oxide nanoparticle-embedded hydrogels enhance nutrient supply and growth of lettuce (Lactuca sativa) infected with Fusarium oxysporum f. sp. lactucae. EnST. 2021;55:13432–13442. doi: 10.1021/acs.est.1c00777. [DOI] [PubMed] [Google Scholar]
- 43.Li M., Lv R., Zhang L., Zi X., Zhou H., et al. Melatonin is a promising silage additive: Evidence from microbiota and metabolites. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.670764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conway J.M., Walton W.G., Salas-González I., Law T.F., Lindberg C.A., et al. Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome. Nat Microbiol. 2022:1–17. doi: 10.1038/s41564-022-01244-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun L., Wang Y., Ma D., Wang L., Zhang X., et al. Differential responses of the rhizosphere microbiome structure and soil metabolites in tea (Camellia sinensis) upon application of cow manure. BMC Microbiol. 2022;22 doi: 10.1186/s12866-022-02470-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haichar Fe.Z., Cernava T., Liu J., Timm C.M. Novel insights into the response of the plant microbiome to abiotic factors. Front Plant Sci. 2021;12 doi: 10.3389/fpls.2021.607874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang X., Liu S., Liu X., Zhang S., Li L., et al. Plant pathological condition is associated with fungal community succession triggered by root exudates in the plant-soil system. Soil Biol Biochem. 2020;151 [Google Scholar]
- 48.Williams A., de Vries F.T. Plant root exudation under drought: implications for ecosystem functioning. New Phytol. 2020;225:1899–1905. doi: 10.1111/nph.16223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material