Highlights
-
•
A novel PCR protocol was developed to detect pkdhps polymorphisms.
-
•
The R34L pkdhfr and quadruple pkdhps mutations are the most prevalent in Thailand.
-
•
Molecular docking showed five pkdhps mutations were less likely to induce drug resistance.
-
•
It was suggested that eastern Thailand's isolates are closely related to Cambodia.
Keywords: Plasmodium knowlesi, pkdhfr, pkdhps
Abstract
Background
The 2022 malaria WHO reported around 4000 P. knowlesi infections in the South-East Asia region. In the same period, 72 positive cases were reported by the Department of Disease Control in Thailand, suggesting a persistent infection. Little is known about dihydrofolate reductase (pkdhfr) and dihydropteroate synthase (pkdhps), putative antimalarial resistance markers for P. knowlesi. The relevant amplification and sequencing protocol are presently unavailable. In this study, we developed a protocol for amplifying and evaluating pkdhps mutations. The haplotype pattern of pkdhfr–pkdhps in Thai isolates was analyzed, and the effects of these pkdhps mutations were predicted by using a computer program.
Methods
Pkdhps were amplified and sequenced from 28 P. knowlesi samples collected in 2008 and 2020 from nine provinces across Thailand. Combining pkdhfr sequencing data from previous work with pkdhps data to analyze polymorphisms of pkdhfr and pkdhps haplotype. Protein modeling and molecular docking were constructed using two inhibitors, sulfadoxine and sulfamethoxazole, and further details were obtained through analyses of protein–ligand interactions by using the Genetic Optimisation for Ligand Docking program. A phylogenetic tree cluster analysis was reconstructed to compare the P. knowlesi Malaysia isolates.
Results
Five nonsynonymous mutations in the pkdhps were detected outside the equivalence of the binding pocket sites to sulfadoxine and sulfamethoxazole, which are at N391S, E421G, I425R, A449S, and N517S. Based on the modeling and molecular docking analyses, the N391S and N517S mutations located close to the enzyme-binding pocket demonstrated a different docking score and protein–ligand interaction in loop 2 of the enzyme. These findings indicated that it was less likely to induce drug resistance. Of the four haplotypes of pkdhfr–pkdhps, the most common one is the R34L pkdhfr mutation and the pkdhps quadruple mutation (GRSS) at E421G, I425R, A449S, and N517S, which were observed in P. knowlesi in southern Thailand (53.57%). Based on the results of neighbor-joining analysis for pkdhfr and pkdhps, the samples isolated from eastern Thailand displayed a close relationship with Cambodia isolates, while southern Thailand isolates showed a long branch separated from the Malaysian isolates.
Conclusions
A new PCR protocol amplification and evaluation of dihydropteroate synthase mutations in Knowlesi (pkdhps) has been developed. The most prevalent pkdhfr-pkdhps haplotypes (53.57%) in southern Thailand are R34L pkdhfr mutation and pkdhps quadruple mutation. Further investigation requires additional phenotypic data from clinical isolates, transgenic lines expressing mutant alleles, or recombinant proteins.
Graphical abstract
This is special type of abstract that is so short and could be inserted after main abstract of article, as a blurb or inserted as annotations into a Table of contents
1. Introduction
With the implementation of a strategy for global malaria elimination by 2030, WHO reported a reduction of 80% and 77% in estimated cases and deaths in Southeast Asia in 2020 compared with 2010. Most reported malaria cases were due to Plasmodium vivax and P. falciparum (WHO, 2021). In 2004, the emergence of human infections with simian malaria (P. knowlesi) was reported in Malaysia Borneo (Singh et al., 2004), which was subsequently reported in other countries of Southeast Asia: Cambodia (Imwong et al., 2019), Indonesia (Figtree et al., 2010), Laos (Iwagami et al., 2018), Myanmar (Jiang et al., 2010), Singapore (Ng et al., 2008), Philippines (Luchavez et al., 2008), Vietnam (Van den Eede et al., 2010), and Thailand (Jongwutiwes et al., 2004). These incidences indicate that human P. knowlesi infections are more prevalent than assumed. Consequently, P. knowlesi is recognized as the fifth human malaria-causing agent that poses a new challenge to the global malaria elimination program (White, 2008). Long- and pig-tailed macaques (Macaca fascicularis and M. nemestrina, respectively) are natural reservoir hosts of P. knowlesi, which limit its distribution in Southeast Asia (Garnham, 1966). The first naturally acquired human P. knowlesi infection from traveling to Malaysia was reported in 1965 (Chin et al., 1965). In Thailand, P. knowlesi infection was reported in a patient who traveled to Prachuap Khiri Khan in 2004 (Jongwutiwes et al., 2004). P. knowlesi infection has also been reported in southern Thailand, where simian parasites are circulating among their natural hosts (Putaporntip et al., 2010; Putaporntip et al., 2021; Sugaram et al., 2021).
The WHO guidelines for malaria 2022 recommend artemisinin-combined therapy or chloroquine (CQ) for uncomplicated P. knowlesi infection and artesunate injection for complicated infection (WHO, 2022). To date, reports of P. knowlesi being resistant to antimalarial drugs have been lacking, and no mutations have been found in the chloroquine resistance transporter orthologous resistance genes of P. falciparum (Tyagi et al., 2013). Additionally, the six clinical isolates evaluated for drug susceptibility were susceptible to CQ and artemisinin and less sensitive to mefloquine (Fatih et al., 2013). Thus far, the evidence indicates that P. knowlesi is sensitive to antimalarial drugs.
Pyrimethamine and sulfadoxine (SDX) inhibit two critical enzymes in the folate biosynthesis pathway of parasites, namely dihydrofolate reductase (DHFR) and dihydropteroate synthase (DHPS). The molecular mechanism underlying antifolate resistance has been associated with a specific point mutation in pf/pvdhfr and pf/pvdhps genes (Korsinczky et al., 2004; Wang et al., 1997). Resistance mechanisms in P. falciparum and P. vivax have been characterized, and mutations in DHFR, and DHPS enzyme-binding pockets led to decreased drug-binding affinity (Chitnumsub et al., 2020; Yogavel et al., 2018; Yuvaniyama et al., 2003). Resistance levels have been associated with the mutation of several positions in each gene. Quadruple pf/pvdhfr mutation resulted in the highest pyrimethamine resistance level. The double mutation A437G-K540E of pfdhps and triple mutation S382A-A383G-A553G of pvdhps were found to be correlated with SDX resistance in P. falciparum and P. vivax (Japrung et al., 2007; Sirawaraporn et al., 1997; Triglia et al., 1997; Yogavel et al., 2018). Both pf/pvdhfr and pf/pvdhps mutations occur sequentially, with the first often causing a mutation in dhfr, followed by dhps when parasite populations carry at least two mutations in dhfr (Imwong et al., 2003; Lozovsky et al., 2009; Plowe et al., 1997).
Mechanisms of antifolate resistance in P. falciparum and P. vivax are well understood, but little is known about pkdhfr and pkdhps genes in P. knowlesi. A few reports are available on polymorphism in pkdhfr, including isolates from India (Tyagi et al., 2013), Malaysia (Grigg et al., 2016), Cambodia (Imwong et al., 2019), and Thailand (Sugaram et al., 2021). By contrast, a recent study reported observing numerous pkdhps polymorphisms in Malaysian isolates (Saif, 2023). In addition, a protocol for pkdhps amplification and sequencing and information regarding the pkdhfr–pkdhps haplotype in P. knowlesi are lacking.
These limitations in pkdhps-related information represent a research gap that needs to be filled to achieve a better understanding of the biology and molecular epidemiology of drug resistance in P. knowlesi malaria parasites. This information can help support eliminate malaria infections in these regions due to the increasing incidence of P. knowlesi. In this study, seminested PCR was performed using newly designed primers to amplify the pkdhps gene. The frequencies and haplotype patterns of pkdhfr–pkdhps in human P. knowlesi infections have been reported in Thailand. The effects of observed pkdhps enzyme-binding site mutations were investigated through protein modeling, molecular docking, and protein interaction. Moreover, phylogenetic trees were constructed using a partial sequence of Thai pkdhfr and pkdhps genes to compare them with published Malaysian isolate data.
2. Material and methods
2.1. Study sites and sample collection
Dried blood spots of 28 symptomatic malaria were collected between 2008 and 2020 from 9 provinces of Thailand, namely Chanthaburi, Chumphon, Phang-nga, Prachuap Khiri Khan, Ranong, Surat Thani, Surin, Trat (Sugaram et al., 2021), and Tak province which included in this study (Table S1). Of these samples, 27 samples were mono-P. knowlesi infections, while one sample from the Tak Province was a mixed infection with P. vivax. All blood samples were extracted using the QIAamp® DNA Mini and Blood Mini kit (Qiagen, Germany) following the manufacturer's instructions.
2.2. Pkdhps amplification and haplotype analysis
To determine gene mutation, primers were designed, the gene was amplified, and sequences were analyzed for the partial pkdhps fragment. The gene locus PKNH_1429900 on NCBI was used as a reference sequence to design primers for pkdhps amplification by using the Primer3Plus program and OligoCalc (Kibbe, 2007; Untergasser et al., 2007). Pkdhps was amplified through seminested PCR and verified through Sanger sequencing (Macrogen, Korea). Although P. knowlesi has 80% nucleotide identity with P. vivax, its cross-reactivity was tested, and no cross-reactivity was observed in the primers. Table 1 presents the primers and PCR conditions used to amplify the gene. The nucleotide and amino acid sequences of these amplicons were confirmed by blasting against the NCBI database. Neutrality selection was calculated using DnaSP software within a sliding window of 100 bp and a step size of 25 bp. MEGA-X software was used to analyze point mutations by comparing the sequences generated with the reference sequences (gene locus: PKNH_1429900) and analyzing haplotypes with pkdhfr data from the previous study (Sugaram et al., 2021) (Table S1).
Table 1.
Specific primers and PCR conditions were used to amplify pkdhps gene.
| Primers | Sequence (5’→3’) | PCR conditions | Product (bp) |
|---|---|---|---|
| PkDHPS_N1F | ATGTTAACTACGATTCCTTTTCTG | nest1: 94°C for 30 s; 50°C for 30 s; 72°C for 1 min for 35 cycles. |
795 |
| PkDHPS_R | ATTTCCCTTTAGTCAGTTCTAGG | ||
| PkDHPS_N2F | TATTCGAAATGACGAATGATGG | nest2: 94°C for 30 s; 55°C for 30 s; 72°C for 1 min for 30 cycles. | 729 |
2.3. Phylogenetic trees analysis
By using a partial 649-bp sequence of pkdhfr from the study samples, Cambodian isolates (Imwong et al., 2019) and Malaysian isolates (Grigg et al., 2016), the neighbor-joining tree was built using the Kimura 2-parameter nucleotide substitution model and gamma distribution (K2+G), the best substitution model based on the Bayesian Information Criterion (BIC). To construct the pkdhps tree, partial 651-bp Thailand and Cambodia that was reported in this study and 28 extracted Malaysia datasets (Table S1) were processed using the Tamura 3-parameter base substitution method (T92+G). The pkdhps datasets from Malaysia were obtained from the primary genome databases. The study conducted by (Saif, 2023), utilized gene polymorphism data obtained from (Assefa et al., 2015; Pinheiro et al., 2015) as the data source to generate polymorphic samples of each Malaysian sample. Thousand bootstrap replications were used to evaluate the trees.
2.4. Protein homology modeling and molecular docking
The 3D structure of the PkDHPS protein was modeled using the SWISS-MODEL server with default settings. The protein sequences were entered in FASTA format, and 3D homology models were retrieved as PDB files and used for molecular docking with its inhibitors.
The 3D structures of the ligands 4-aminobenzoic acid (pABA), SDX, and sulfamethoxazole (SMZ) were loaded from PubChem. These ligands were used for protein docking by using the Genetic Optimisation for Ligand Docking program. Each ligand was docked within 10 Å of protein-binding site atoms and run with a genetic algorithm 100 times without the early termination. The best docking poses were selected based on the highest ASP scoring function. BIOVIA Discovery Studio Visualizer V21.1.0 analyzed the interaction of the best pose complexes. All protein structures and complexes were compared and visualized using the PYMOL Molecular Graphics System, V2.5.2.
3. Results
3.1. Polymorphism of pkdhfr–pkdhps in human isolates from Thailand
The partial pkdhps of 729 bp (codon 364th–606th of pkhppk-dhps) was successfully amplified and sequenced from 28 patients in Thailand. Eleven polymorphisms were found, which consisted of 6 synonymous and 5 nonsynonymous mutations (Fig. 1). However, these nonsynonymous mutations were not observed in the five residues of the protein–ligand binding pocket of PkDHPS (namely S382, A383, K514, A555, and V587), which are equivalent to PvDHPS and PfDHPS that correspond to sulfa drug resistance (Table S2). A silent mutation was observed at the encoded S382 codon, which altered the third nucleotide from TCC to TCT (17/28), 60.71%, and is mainly found in the sample from southern Thailand.
Fig. 1.
The frequency of PkDHPS mutations from human P. knowlesi infection in Thailand and the five residues (S382, A383, K514, A555, and V587) in PkDHPS that correspond to the sulfadoxine binding sites of PvDHPS (S382, A383, K512, A553, V585) and PfDHPS (S436, A437, K540, A581, A613) are shown.
Five point mutations were detected outside the enzyme-binding pocket in 18 of the 28 pkdhps. A quadruple mutation (GRSS) in residues E421G, I425R, A449S, and N517S was detected in isolates from southern Thailand (53.57%; 15/28): Chumphon (10/28), Ranong (1/28), Phang-nga (2/28), Surat Thani (1/8), and Prachuap Khiri Khan (1/28). The triple mutation (GRS) in the aforementioned residues, was found in 1 isolate (7.14%) each from Chumphon and Prachuap Khiri Khan. By contrast, 10 samples from the Thai-Cambodian border (35.71%; 10/28), including Surin (1/28), Trat (8/28), and Chanthaburi (1/28), were wildtypes (Table 2; Fig. S1). Furthermore, 1 sample from Tak Province had coinfection with P. vivax and revealed a mixed genotype of pkdhps at I425I/R and a unique mutation at N391S.
On combining pkdhps and pkdhfr from the previous report, four haplotype patterns appeared among the 28 clinical Thai isolates, as shown in Table 2. The two most prevalent haplotypes were haplotypes 1 (53.57%; 15/28) and 3 (35.71%; 10/28), respectively. Haplotype 1 consisted of a single R34L dhfr, and a quadruple dhps mutation was found in samples collected from southern Thailand provinces. While Haplotype 3 carried T105 deletion dhfr, a wildtype pkdhps sequence was noted in samples from provinces along the Thai-Cambodia border (Fig. S1). Four haplotypes within a nucleotide diversity (π) of 0.007 were identified across the pkdhps sequences of Thailand isolates, indicating the genetic diversity of Thailand isolates. A neutrality selection test statistic was observed non-significantly positive selection of Tajima's D (1.962), Fu and Li's D (0.457), and F (1.072) at p-value > 0.10 (Fig. S2).
Table 2.
The pkdhfr-pkdhps haplotypes of P. knowlesi isolates in Thailand. Bold letters indicate the amino acid substitution and deletion mutation.
| DHFR(Sugaram et al., 2021) | DHPS | Frequency (n=28) | Study site | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P. falciparuma strain 3D7 | V5 | Y35 | L81 | K96 | K445 | - | - | A475 | N543 | ||
| P. vivaxb strain SalI | L4 | R34 | L80 | D105 | S391 | V421 | A425 | A447 | N515 | ||
| P. knowlesi | |||||||||||
| Wildtypec | L4 | R34 | L80 | T105 | N391 | E421 | I425 | A449 | N517 | 0% (0/28) | |
| Haplotype 1 | L | L | L | T | N | G | R | S | S | 53.57% (15/28) | Phang-nga, Ranong, Surat Thani, Chumphon, Prachuap Khiri Khan |
| Haplotype 2 | L | L | L | T | N | G | R | S | N | 7.14% (2/28) | Chumphon, Prachuap Khiri Khan |
| Haplotype 3 | L | R | L | Del | N | E | I | A | N | 35.71% (10/28) | Chanthaburi, Trat, Surin |
| Haplotype 4 | F | R | L | T | S | E | I/R | S | N | 3.57% (1/28d) | Tak |
Gene locus of P. falciparum: pfdhfr (PF3D7_0417200), pfdhps (PF3D7_0810800).
Gene locus of P. vivax: pvdhfr (PVX_089950), pvdhps (PVX_123230).
Alignment against P. knowlesi strain H: pkdhfr (PKNH_0509600), pkdhps (PKNH_1429900).
P. knowlesi co-infected with P. vivax.
3.2. Homology PkDHPS protein modeling
The effect of mutations on the inhibitor-binding sites of SDX and SMZ was determined through protein homology modeling and docking. Each point mutation N391S, E421G, I425R, A449S, and N517S was built separately into a protein model for further analysis. The built PkDHPS models were based on the PvDHPS crystal structure (PDB ID: 5z79) with 80.45%–80.73% sequence identity and covered 95% of the pkdhps sequence (Yogavel et al., 2018). Loop structure of building structures at positions 590–697 of P. knowlesi were disordered crystal structures of PvDHPS. In addition, these P. knowlesi residues had two insertion sites at residues 599–626 and 675–681.
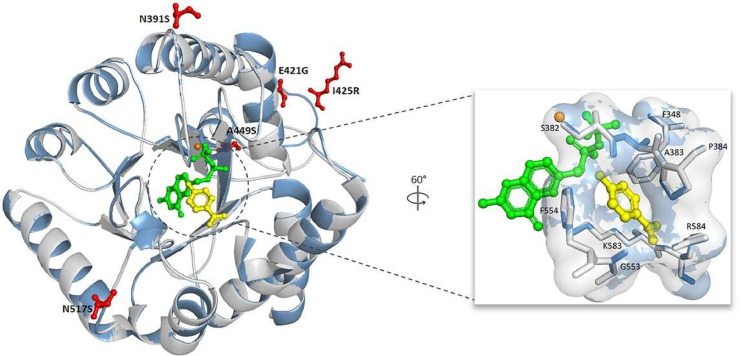
Among the five mutations of PkDHPS, N391S, and N517S was part of the active site loop 2 and loop 5 of the enzyme, according to structural alignment against the PfDHPS and PvDHPS structures. N391S, located on loop 2 is involved in the flexibility conformation of the loop, causing a change in ligand–protein affinity. N517S was close to loop 5, forming a salt-bridge gate with loop 7 (Fig. 2) (Chitnumsub et al., 2020; Yogavel et al., 2018). Hence, mutants carrying these mutations were used to further characterize the effect of the inhibitor-binding affinity through protein docking.
Fig. 2.
The homology model of PkDHPS was constructed, and the superposition of the wildtype (light gray) and mutant PkDHPS (blue) structures was annotated. It included a magnified view of the active site and the identification of contact residues, which were displayed within a compact enclosure. Furthermore, the mutations identified in this investigation are visually represented as a red-colored ball and stick model. The bound substrates pABA (yellow) and PtPP (green) are depicted in a ball and stick representation, while the cofactor magnesium is represented by an orange sphere.
3.3. Effect of mutations in PkDHPS protein on inhibitor binding
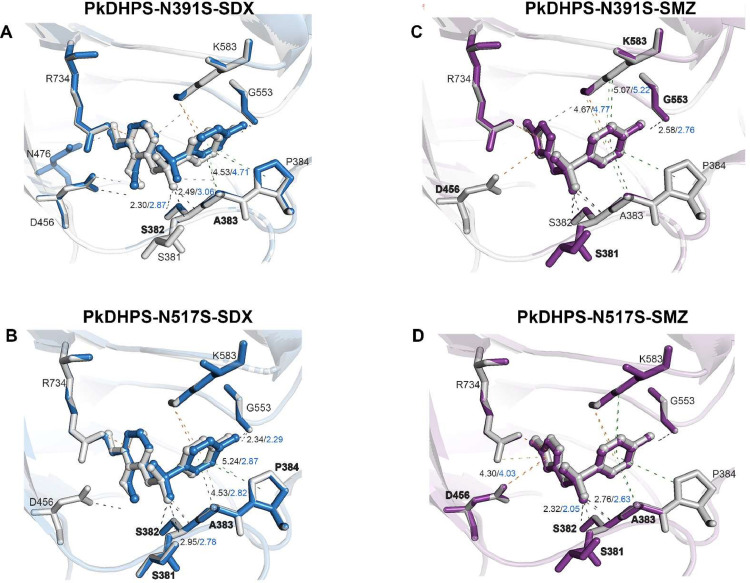
Only two residues, N391S, and N517S, were more closely located near the active site enzyme. We further characterized the effect of mutations on inhibitor binding through molecular docking within 10 Å of the active site and ran 100 times. The three ligands used in this study were the natural substrate pABA and two inhibitors, SDX, and SMZ. The docking result revealed that the binding score of the N391S PkDHPS mutant and ligands decreased, whereas that of the N517S mutant and ligands increased compared to the wild type (Table 3). The interaction of contact residues influences the difference in the docking score of P. knowlesi mutants. A loss of putative hydrogen bonds from S381 to SDX was observed in the N391S mutant. Moreover, the contact distance between residues S382, A383, and SDX increased compared to wild-type interactions, thereby reducing the docking score. By contrast, the N517S mutant revealed a reduced contact distance between SDX and residues S381, S382, A383, and P384 of PkDHPS, thereby causing an increase in the docking score (Fig. 3A-B).
Table 3.
ASP scoring function of PkDHPS inhibitors and their natural substrates.
| Variants | pABA | Sulfadoxine (SDX) | Sulfamethoxazole (SMZ) |
|---|---|---|---|
| Wildtype P. knowlesi | 24.23 | 35.35 | 31.88 |
| Mutant P. knowlesi | |||
| N391S | 24.10 (0.13) | 34.42 (0.93) | 31.71 (0.17) |
| N517S | 24.31 (-0.08) | 36.03 (-0.68) | 31.98 (-0.10) |
Note: ASP scores have no unit and refer to the fitness of ligands. A higher score indicated a high affinity of ligands for the protein. The scores in the bracket refer to the relative docking scores.
Fig. 3.
The interaction between PkDHPS, sulfadoxine (SDX), and sulfamethoxazole (SMZ) has been observed. The structural components representing the interaction residues between the wildtype (gray) and mutant variants (blue; SDX; purple; SMZ) were visualized in the form of stick representations. The dashed line in the diagram represents three types of interactions: hydrogen bonds (gray), hydrophobic interactions (green), and electrostatic interactions (orange). The lighter colors within these interactions indicate interactions involving mutants. The distances between ligands and contact residues are represented by black and blue colors for the wildtype and mutant samples, respectively. The differences in interaction and distance residues between the wild-type and mutant variants are clearly indicated.
In the PkDHPS-SMZ complexes, the unique interaction of the N391S mutant with residues S381 and K583, involves hydrogen bonds, whereas the wildtype exhibits pi-cation interaction with D456. Furthermore, the distance slightly increased between the inhibitor and mutant protein residues G553 and K583 compared to the wild type. The interaction of the N517S mutant also exhibited a unique hydrogen bond interaction with residue S381. The contraction of distance between residues S382, A383, and D456 of the N517S mutant and SMZ could lead the inhibitor closer to the protein and more tight binding, resulting in an increased docking score (Fig. 3C-D). However, further experiments are required to investigate whether these mutations affect protein–inhibitor binding.
3.4. Phylogenetic tree analysis based on pkdhfr and pkdhps genes
Phylogenetic tree cluster analysis was reconstructed to compare P. knowlesi from Thailand and Malaysian isolates from previous studies based on pkdhfr (Grigg et al., 2016) and pkdhps ( Saif, 2023) genes. The neighbor-joining tree is presented in Fig. 4, Fig. 5. Thailand samples isolated from Chanthaburi, Surin, and Trat were closely related to Cambodian isolates and were supported with a reliable bootstrap of 77% and 89% of pkdhfr and pkdhps, respectively, which were also observed in a previous pkmsp1 analysis (Sugaram et al., 2021). By contrast, the southern Thailand isolates had a long branch separated from that of the Malaysian isolates, with 67% and 72% bootstrap reliability, and formed monomorphic clustering, except for 1 sample from Chumphon and Prachuap Khiri Khan provinces in the pkdhps tree. The sample from Tak Province was positioned differently from the clade of Thailand samples, consistent with a unique haplotype pattern. Additionally, the trees revealed sequence diversity in both genes among human Malaysian isolates.
Fig. 4.
The neighbor-joining tree was constructed based on 649 bp of pkdhfr gene from P. knowlesi isolates in this study (Thailand ▲), Cambodia (■) (Imwong et al., 2019) and Malaysia (●) (Grigg et al., 2016). The colors are defined for each mutation position on the pkdhfr gene. The tree was supported by 1000 bootstraps. Red asterisk (*) indicated the P. vivax mixed infection isolate.
Fig. 5.
The neighbor-joining tree of the pkdhps gene, from samples collected in Thailand (▲), Cambodia (■), and Malaysia (●) (Saif, 2023), was constructed using a bootstrap analysis with 1000 replicates. The markers that were colored indicated the presence of a nonsynonymous mutation on the gene. A representative specimen pertaining to the M. nemestrina host was highlighted with a light green color. Red asterisk (*) indicated the P. vivax mixed infection isolates.
4. Discussion
The National Malaria Elimination Strategy Thailand, 2017–2026, has been implemented with the vision that Thailand will be malaria-free in 2024 (Bureau of Vector Borne Diseases, 2016). Global malaria cases were reduced from 48,109 to 24,850 from 2012 to 2015 (WHO, 2021). In Thailand, malaria cases reduced from 5,432 to 3,266 between 2019 and 2021. Between 2021 and 2022, simian malaria P. knowlesi infection had exponentially increased from 72 to 176 cases during the COVID-19 pandemic, creating a barrier for public health staff entering the endemic areas to monitor and evaluate the disease status, including making it inconvenient for patients to enter the public health care system. Moreover, in the first quarter of 2023, the Department of Disease Control of Thailand, reported 2,103 positive malaria cases in Thailand, with P. knowlesi being the top two in terms of causing the highest number of infections (105/2103). The majority of the cases 92.38% (97/105) were reported from southern Thailand provinces (Division of Vector Borne, 2020). This was a 59.66% increase from positive malaria cases in 2022, indicating an increasing incidence of P. knowlesi infection in Thailand. Thus, this species should be acknowledged as one that needs to be controlled and eliminated from Thailand. Studies have found that P. knowlesi infection predominates in southern Thailand (Putaporntip et al., 2021; Sermwittayawong et al., 2012; Sugaram et al., 2021), which is close to Malaysia, where positive P. knowlesi infection has been predominant in humans and a high prevalence of P. knowlesi has also been reported in natural hosts (Akter et al., 2015; Lee et al., 2011; Saleh Huddin et al., 2019) Thus, the infection could have been transmitted to Thailand via the migration of hosts and vectors.
Attempts to study antimalarial resistance in antimalarial resistance markers of P. knowlesi samples have been reported in pkcrt (Fatih et al., 2013; Tyagi et al., 2013), pkdhfr (Grigg et al., 2016; Imwong et al., 2019; Sugaram et al., 2021; Tyagi et al., 2013), pkkelch, and pkdhps (Saif, 2023) genes, which are ortholog resistance genes in P. falciparum and P. vivax. A polymorphic pkdhfr, but not pkdhps, has been reported in Thailand (Sugaram et al., 2021). Moreover, a simple protocol to screen for pkdhps mutation is lacking. Thus, this study provided preliminary data on the status of antifolate resistance and a haplotype pattern of pkdhfr–pkdhps genes in P. knowlesi from Thailand by using validated PCR protocols for pkdhps point mutation screening. The study samples were limited because a low number of positive P. knowlesi infections had been detected in Thailand in the collection year 2019–2020 (Division of Vector Borne, 2020). Additional information from Thailand and other regions will be required to fill in all the gaps.
Five nonsynonymous mutations, N391S, E421G, I425R, A449S, and N517S, which were outside the PkDHPS enzyme-binding site, were detected in this study. This finding indicated that P. knowlesi has remained susceptible to antifolate drugs and has not undergone drug selection in the parasite population in Thailand. The absence of a positive selection signal on pkdhps, consistent with the whole-genome sequencing results in Malaysia, indicates that its reservoir hosts have never been exposed to drug pressure, and so, no mutation exists on this resistance marker (Assefa et al., 2015). These mutations were primarily observed in southern Thailand, whereas samples from provinces along the Thai-Cambodia border contained wild-type sequences throughout the gene. A recent study revealed 73 polymorphisms throughout the pkhppk-dhps encoding bifunctional protein of P. knowlesi in Malaysia. Of them, 20 polymorphisms were in the residues equivalent to those observed in this study (13 synonymous and 7 nonsynonymous mutations) (Saif, 2023), except the I425R mutation, which was observed in both studies. The majority of Malaysia's genotypic pattern was wild type (19/28, 67.86%) (Table S4). Therefore, the pkdhps genotype of P. knowlesi in Thailand differed from that in Malaysia. Interestingly, the I425R mutation was detected at distinct frequencies in Thailand and Malaysia samples; however, a higher prevalence was observed in samples from southern Thailand (60.71 %; 17/28) than in samples from Malaysia (7.14 %; 2/28) (Saif, 2023). Furthermore, a single sample from Tak Province, northern Thailand, revealed a distinct pkdhps genotype with a mutation at residue N391S and a mixed genotype at I425I/R. Therefore, additional samples from northern Thailand should be collected to investigate this unique finding.
The pvdhfr quadruple mutation LRMT (F57L-S58R-T61M-S117T) increased the IC50 of pyrimethamine resistance by approximately 500-fold (Hastings et al., 2004), and the pvdhps double mutation GG (A383G-A553G) was observed to exhibit high-level SDX resistance compared to the wildtype (Yogavel et al., 2018). Combining pvdhfr and pvdhps mutations with sextuple mutations resulted in high-grade resistance, which resulted in clinical treatment failure in P. vivax infection (Imwong et al., 2005). In this and other studies, no mutations were observed in the predicted binding pocket. The resistance level of P. knowlesi was analyzed for the enzyme PkDHFR but not PkDHPS. At least a nanomolar concentration of pyrimethamine inhibited PkDHFR, significantly influencing the schizont stage of the parasite's lifecycle (Gutteridge & Trigg, 1971). Moreover, one isolate from southern Thailand was recently characterized by an inhibitory constant (Ki) that was more sensitive to pyrimethamine than the wild type of other Plasmodium species (Ittarat et al., 2018), which is consistent with in vitro pyrimethamine testing that P. knowlesi revealed 10-fold drug susceptibility than P. falciparum (van Schalkwyk et al., 2017). Furthermore, in vitro testing of PkDHFR inhibitors had been reported in clinical isolated from Malaysia that showed similar drug susceptibility to the lab adapted strain (van Schalkwyk et al., 2019). Additional in vitro and in vivo investigations of PkDHPS requires additional research.
This study identified two pkdhfr mutations located outside the enzyme-binding site (Sugaram et al., 2021), namely R34L substitution (17/28; 60.71%) and T105 deletion (10/28; 35.71%), in Thai isolates. The R34L mutation was the predominant genotype detected in Thailand. It was found in samples from southern Thailand that border Malaysia, where the mutation detection rate was 8.91% (40/449), which was among the top three highest mutation detection rates in Sabah, Malaysia (Grigg et al., 2016). By contrast, the T105 deletion was identified in provinces along the Thai-Cambodia border, corresponding to data from pkdhfr from Cambodia's Pailin and Battambang provinces, but has never been reported from Malaysia (Table S3). This deletion, however, did not affect the pkdhfr frameshift (Imwong et al., 2019). Previous protein modeling studies have reported that the R34L mutation and T105 deletion were not related to drug resistance in P. knowlesi because the positions were far from the enzyme's binding site (Grigg et al., 2016; Sugaram et al., 2021). In addition, a novel independent mutation at L4F was identified in one sample from Tak Province, a border province between Thailand and Myanmar.
Finding the pkdhps mutation allowed us to identify pkdhfr and pkdhps haplotypes in Thai isolates and observe four haplotypes in our investigation. Over half of the parasites in Thailand were Haplotype 1, with the pkdhfr single mutation (R34L) combined with the pkdhps quadruple mutation (E421G, I425R, A449S, and N517S). Those samples were exclusively from southern Thailand, whereas the sample from the Thai-Cambodia border regions was Haplotype 3 (35.71%; 10/28), indicating a pkdhfr T105 deletion and the pkdhps wildtype sequence. Thus, the findings indicate that P. knowlesi haplotype patterns differ between geographic locations. This highest prevalence of Haplotype 1 in Thailand could be concerning, and further investigation is required to prove whether a correlation exists between these mutations and antifolate resistance in P. knowlesi in that part of Thailand.
In addition, the effect of quintuple mutations in pkdhps was evaluated through protein modeling, protein docking, and interaction analysis with PkDHPS. The protein homology of PkDHPS was derived on the basis of the PvDHPS (PDB ID: 5z79) crystal structure and shared sequences with approximately 80% degree of similarity. Nonetheless, the PkDHPS model identified loop structure at positions 590–697, which corresponded to the disordered PvDHPS structure that may play a role in protein interface dimerization, but is unrelated to the binding of substrate or inhibitors, as reported in the PfDHPS crystal structure (Chitnumsub et al., 2020; Yogavel et al., 2018). Differences in hydrogen bond formation and the contact distance between loop 2 (S381, S382, A383, and P384) residues of protein and the drug led to differences in the docking score of the complex of SDX with N391S and N517S mutants as well as of the complex of N517S mutant and SMZ. The distinct contact distances of the SMZ–N391S mutant at G553 and K583 were found on loops 6 and loop 7, respectively, of the binding pocket. This finding suggests that the interaction in loop 2 of the protein binding pocket had a greater impact on differences in the docking score.
The PvDHPS and PfDHPS crystal structures revealed that loop 2 is a flexible loop having various conformations depending on the bound ligand, which results in different ligand affinities (Chitnumsub et al., 2020; Yogavel et al., 2018). In a previous study, PfDHPS residues S436, A437, and A613 were located within 3 Å of the binding site, causing a direct effect on SDX binding. Using the enzyme inhibition assays in PfDHPS (S436, A437) and PvDHPS (S382, A383), the critical residues of loop 2 were determined. The S436A and A437G double mutations of PfDHPS resulted in a 13-fold decrease in SDX binding compared to the wild type (Chitnumsub et al., 2020), while the A383G mutation of PvDHPS led to a 47-fold decrease (Pornthanakasem et al., 2016). However, the interaction between wild-type and mutant organisms revealed minor differences in this study. While computational predictions can provide valuable insights into the prediction of drug resistance mutations, they may also exhibit limitations in accurately identifying such alterations. To clarify the direct influence of these mutations on drug resistance, the inclusion of phenotypic data obtained from clinical isolates or experimental outcomes is crucial in order to validate the accuracy and dependability of these predictions.
In this study, dendrograms of Thai pkdhfr, and pkdhps demonstrated differentiated sample clusters between Peninsular and Borneo Malaysia, which is consistent with a previous finding on the single-nucleotide polymorphism (SNP) tree from whole-genome sequencing (Assefa et al., 2015). The Peninsular clade comprises most Thai isolates in the pkdhfr tree, but the southern Thailand isolates were assigned to a different clade in the pkdhps tree. Samples from the Thai-Cambodian borders shared similar genotypes with those from Cambodia, which aggregated together in the same taxa in both trees. This indicated that the migration of humans or reservoir hosts has resulted in a shared parasite population across these regions. By contrast, the southern Thailand isolates had a long branch from the Malaysian isolates, indicating that P. knowlesi from Malaysia could have spread to southern Thailand, which still retained ancestor characteristics and underwent little evolution. Despite this, most Thailand isolates were monomorphic, suggesting an extended migration of the parasite from other countries, which then spread in Thailand. Individual parasite population expansion in Malaysia is supported by high average nucleotide diversity in the population and the presence of individual taxa in the SNP neighbor-joining tree.
Similar to the previous report (Diez Benavente et al., 2017), the pkdhps tree in this study created sample clusters based on its natural hosts M. fascicularis (Mf-Pk) and M. nemestrina (Mn-Pk). However, the four Mf-Pk samples found in the Mn-Pk taxa and three of the four isolates reported genetic exchange between host-associated clusters at chromosomes 5, 8, and 11 but not at the pkdhps gene on chromosome 14 (Turkiewicz et al., 2023). Thus, this observation would not be a result of the parasite's genetic introgression, but rather due to the information provided by the partial gene analysis, which may be insufficient to distinguish between the two clusters. To confirm this evidence, the entire pkdhps gene must be analyzed and samples from Malaysia and Thailand must be compared. By contrast, host-related samples could not be indicated in the pkdhfr tree (Grigg et al., 2016) owing to a lack of information regarding the host association for the Malaysian isolates. To understand the fundamentals of parasite transmission from macaques to humans, the P. knowlesi structure in Thailand needs to be investigated to determine the relationship between the parasite's natural host and the samples.
This study fills the malaria research gap by developing and providing protocols and primers to screen pkdhps mutations. Moreover, the different pkdhps genotype frequencies between Thailand and Malaysian isolates were revealed, and this was the first study reporting the haplotype pattern of pkdhfr–pkdhps in samples from Thailand. The mutations outside the enzyme-binding pockets of PkDHPS were analyzed. However, further experiments are required to investigate the effect of these mutations on antifolate drug resistance in P. knowlesi. However, the mutation status of other drug-resistance markers should be continually monitored. This would help malaria control programs and could limit the spread of emerging resistance parasites.
5. Conclusions
A novel PCR protocol was developed to amplify and evaluate mutations in dihydropteroate synthase in Knowlesi (pkdhps) that will be useful to fill a malaria research gap. The R34L pkdhfr mutation and pkdhps quadruple mutation are the most prevalent pkdhfr-pkdhps haplotypes (53.57%) in southern Thailand. Based on modeling and molecular docking, P. knowlesi isolates with five nonsynonymous pkdhps mutations were less likely to cause drug resistance. According to a neighbor-joining analysis, eastern Thailand isolates are closely related to Cambodia, whereas southern Thailand isolates have a long branch. Additional phenotypic data from clinical isolates, transgenic lines expressing, or recombinant proteins in the mutant alleles are required for further investigation.
CRediT authorship contribution statement
Raweewan Sangsri: Methodology, Validation, Software, Data curation, Formal analysis, Visualization, Writing – original draft. Kiattawee Choowongkomon: Software, Formal analysis. Runch Tuntipaiboontana: Software, Formal analysis. Rungniran Sugaram: Resources. Patcharida Boondej: Resources. Prayuth Sudathip: Resources. Arjen M. Dondorp: Funding acquisition, Writing – review & editing. Mallika Imwong: Conceptualization, Investigation, Supervision, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Ethical approval
Ethical approval for the study was obtained from the ethical review committees of Faculty of Tropical Medicine, Mahidol University (MUTM2020-082-03 and MUTM2023-015-01).
Acknowledgements
We thank Ms. Watcharee Pagornrat, Ms. Wanassanan Madmanee, Ms. Kanokon Suwannasin, and Ms. Jindarat Kouhathog for their help throughout the project.
Funding
This research project was supported by Mahidol University, Fundamental Fund: fiscal year 2023 by National Science Research and Innovation Fund (NSRF) to MI, and part of the Mahidol-University Oxford Tropical Medicine Research Programme funded by the Wellcome Trust of the UK (core grant 106698/B/14/Z) and Wellcome OA statement. This research was funded in whole, or in part, by the Wellcome Trust [220211]. For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. The funding sources did not participate in data analysis or the final decision to publish the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.actatropica.2023.107016.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- Akter R., Vythilingam I., Khaw L.T., Qvist R., Lim Y.A., Sitam F.T., et al. Simian malaria in wild macaques: first report from Hulu Selangor district, Selangor, Malaysia. Malar. J. 2015;14(386) doi: 10.1186/s12936-015-0856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assefa S., Lim C., Preston M.D., Duffy C.W., Nair M.B., Adroub S.A., et al. Population genomic structure and adaptation in the zoonotic malaria parasite Plasmodium knowlesi. Proc. Natl. Acad. Sci. U. S. A. 2015;112:13027–13032. doi: 10.1073/pnas.1509534112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau of Vector Borne Diseases . Ministry of Public Health; Thailand: 2016. National Malaria Elimination Strategy, Thailand 2017–2026. Nonthaburi. [Google Scholar]
- Chin W., Contacos P.G., Coatney G.R., Kimball H.R. A naturally acquited quotidian-type malaria in man transferable to monkeys. Science. 1965;149(865) doi: 10.1126/science.149.3686.865. [DOI] [PubMed] [Google Scholar]
- Chitnumsub P., Jaruwat A., Talawanich Y., Noytanom K., Liwnaree B., Poen S., et al. The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance. FEBS J. 2020;287:3273–3297. doi: 10.1111/febs.15196. [DOI] [PubMed] [Google Scholar]
- Diez Benavente E., Florez de Sessions P., Moon R.W., Holder A.A., Blackman M.J., Roper C., et al. Analysis of nuclear and organellar genomes of Plasmodium knowlesi in humans reveals ancient population structure and recent recombination among host-specific subpopulations. PLos Genet. 2017;13 doi: 10.1371/journal.pgen.1007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Division of Vector Borne Diseases. 2020. Malaria Online. Retrieved 16 March 2023, from Ministry of Public Health, Thailand http://malaria.ddc.moph.go.th/malariar10/index_newversion.php.
- Fatih F.A., Staines H.M., Siner A., Ahmed M.A., Woon L.C., Pasini E.M., et al. Susceptibility of human Plasmodium knowlesi infections to anti-malarials. Malar. J. 2013;12(425) doi: 10.1186/1475-2875-12-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figtree M., Lee R., Bain L., Kennedy T., Mackertich S., Urban M., et al. Plasmodium knowlesi in human, Indonesian Borneo. Emerg. Infect. Dis. 2010;16:672–674. doi: 10.3201/eid1604.091624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham P. Blackwell Scientific Publications Ltd.; Oxford, UK: 1966. Malaria Parasites and Other Haemosporidia. [Google Scholar]
- Grigg M.J., Barber B.E., Marfurt J., Imwong M., William T., Bird E., et al. Dihydrofolate-reductase mutations in Plasmodium knowlesi appear unrelated to selective drug pressure from putative human-to-human transmission in Sabah, Malaysia. PLoS One. 2016;11 doi: 10.1371/journal.pone.0149519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge W.E., Trigg P.I. Action of pyrimethamine and related drugs against Plasmodium knowlesi in vitro. Parasitology. 1971;62:431–444. doi: 10.1017/S0031182000077581. [DOI] [PubMed] [Google Scholar]
- Hastings M.D., Porter Km Fau - Maguire J.D., Maguire Jd Fau - Susanti I., Susanti I Fau - Kania W., Kania W, Fau - Bangs M.J., Bangs Mj Fau - Sibley C.H., et al. Dihydrofolate reductase mutations in Plasmodium vivax from Indonesia and therapeutic response to sulfadoxine plus pyrimethamine. J. Infect. Dis. 2004 doi: 10.1086/381397. [DOI] [PubMed] [Google Scholar]
- Imwong M., Madmanee W., Suwannasin K., Kunasol C., Peto T.J., Tripura R., et al. Asymptomatic Natural Human Infections With the Simian Malaria Parasites Plasmodium cynomolgi and Plasmodium knowlesi. J. Infect. Dis. 2019;219:695–702. doi: 10.1093/infdis/jiy519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M., Pukrittayakamee S., Cheng Q., Moore C., Looareesuwan S., Snounou G., et al. Limited polymorphism in the dihydropteroate synthetase gene (dhps) of Plasmodium vivax isolates from Thailand. Antimicrob. Agents Chemother. 2005;49:4393–4395. doi: 10.1128/AAC.49.10.4393-4395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M., Pukrittayakamee S., Renia L., Letourneur F., Charlieu J.P., Leartsakulpanich U., et al. Novel point mutations in the dihydrofolate reductase gene of Plasmodium vivax: evidence for sequential selection by drug pressure. Antimicrob. Agents Chemother. 2003;47:1514–1521. doi: 10.1128/AAC.47.5.1514-1521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittarat W., Pornthanakasem W., Mungthin M., Suwandittakul N., Leelayoova S., Tarnchompoo B., et al. Characterization of Plasmodium knowlesi dihydrofolate reductase-thymidylate synthase and sensitivity to antifolates. Parasitol. Int. 2018;67:787–792. doi: 10.1016/j.parint.2018.08.004. [DOI] [PubMed] [Google Scholar]
- Iwagami M., Nakatsu M., Khattignavong P., Soundala P., Lorphachan L., Keomalaphet S., et al. First case of human infection with Plasmodium knowlesi in Laos. PLoS Negl. Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japrung D., Leartsakulpanich U., Chusacultanachai S., Yuthavong Y. Conflicting requirements of Plasmodium falciparum dihydrofolate reductase mutations conferring resistance to pyrimethamine-WR99210 combination. Antimicrob. Agents Chemother. 2007;51:4356–4360. doi: 10.1128/AAC.00577-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Chang Q., Sun X., Lu H., Yin J., Zhang Z., et al. Co-infections with Plasmodium knowlesi and other malaria parasites. Myanmar. Emerg. Infect. Dis. 2010;16:1476–1478. doi: 10.3201/eid1609.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongwutiwes S., Putaporntip C., Iwasaki T., Sata T., Kanbara H. Naturally acquired Plasmodium knowlesi malaria in human. Thailand. Emerg. Infect. Dis. 2004;10:2211–2213. doi: 10.3201/eid1012.040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibbe W.A. OligoCalc: an online oligonucleotide properties calculator. Nucleic. Acids. Res. 2007 doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsinczky M., Fischer K., Chen N., Baker J., Rieckmann K., Cheng Q. Sulfadoxine resistance in Plasmodium vivax is associated with a specific amino acid in dihydropteroate synthase at the putative sulfadoxine-binding site. Antimicrob. Agents Chemother. 2004;48:2214–2222. doi: 10.1128/AAC.48.6.2214-2222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.S., Divis P.C., Zakaria S.K., Matusop A., Julin R.A., Conway D.J., et al. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozovsky E.R., Chookajorn T., Brown K.M., Imwong M., Shaw P.J., Kamchonwongpaisan S., et al. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12025–12030. doi: 10.1073/pnas.0905922106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchavez J., Espino F., Curameng P., Espina R., Bell D., Chiodini P., et al. Human Infections with Plasmodium knowlesi, the Philippines. Emerg. Infect. Dis. 2008;14:811–813. doi: 10.3201/eid1405.071407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng O.T., Ooi E.E., Lee C.C., Lee P.J., Ng L.C., Pei S.W., et al. Naturally acquired human Plasmodium knowlesi infection, Singapore. Emerg. Infect. Dis. 2008;14:814–816. doi: 10.3201/eid1405.070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro M.M., Ahmed M.A., Millar S.B., Sanderson T., Otto T.D., Lu W.C., et al. Plasmodium knowlesi genome sequences from clinical isolates reveal extensive genomic dimorphism. PLoS One. 2015;10 doi: 10.1371/journal.pone.0121303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowe C.V., Cortese J.F., Djimde A., Nwanyanwu O.C., Watkins W.M., Winstanley P.A., et al. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J. Infect. Dis. 1997;176:1590–1596. doi: 10.1086/514159. [DOI] [PubMed] [Google Scholar]
- Pornthanakasem W., Riangrungroj P., Chitnumsub P., Ittarat W., Kongkasuriyachai D., Uthaipibull C., et al. Role of Plasmodium vivax dihydropteroate synthase polymorphisms in sulfa drug resistance. Antimicrob. Agents Chemother. 2016;60:4453–4463. doi: 10.1128/AAC.01835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putaporntip C., Jongwutiwes S., Thongaree S., Seethamchai S., Grynberg P., Hughes A.L. Ecology of malaria parasites infecting Southeast Asian macaques: evidence from cytochrome b sequences. Mol. Ecol. 2010;19:3466–3476. doi: 10.1111/j.1365-294X.2010.04756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putaporntip C., Kuamsab N., Seethamchai S., Pattanawong U., Rojrung R., Yanmanee S., et al. Cryptic Plasmodium inui and P. fieldi infections among symptomatic malaria patients in Thailand. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab1060. [DOI] [PubMed] [Google Scholar]
- Saif A. Mutations in Plasmodium knowlesi Kelch protein 13 and the dihydropteroate synthase gene in clinical samples. Asian Pac. J. Trop. Biomed. 2023;16:72–79. doi: 10.4103/1995-7645.370146. [DOI] [Google Scholar]
- Saleh Huddin A., Md Yusuf N., Razak M., Ogu Salim N., Hisam S. Genetic diversity of Plasmodium knowlesi among human and long-tailed macaque populations in Peninsular Malaysia: The utility of microsatellite markers. Infect. Genet. Evol. 2019;75 doi: 10.1016/j.meegid.2019.103952. [DOI] [PubMed] [Google Scholar]
- Sermwittayawong N., Singh B., Nishibuchi M., Sawangjaroen N., Vuddhakul V. Human Plasmodium knowlesi infection in Ranong province, southwestern border of Thailand. Malar. J. 2012;11(36) doi: 10.1186/1475-2875-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B., Kim Sung L., Matusop A., Radhakrishnan A., Shamsul S.S., Cox-Singh J., et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- Sirawaraporn W., Sathitkul T., Sirawaraporn R., Yuthavong Y., Santi D.V. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1124–1129. doi: 10.1073/pnas.94.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugaram R., Boondej P., Srisutham S., Kunasol C., Pagornrat W., Boonyuen U., et al. Genetic population of Plasmodium knowlesi during pre-malaria elimination in Thailand. Malar. J. 2021;20(454) doi: 10.1186/s12936-021-03990-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T., Menting Jg, Fau - Wilson C., Wilson C, Fau - Cowman A.F., Cowman A.F. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 1997 doi: 10.1073/pnas.94.25.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkiewicz A., Manko E., Oresegun D.R., Nolder D., Spadar A., Sutherland C.J., et al. Population genetic analysis of Plasmodium knowlesi reveals differential selection and exchange events between Borneo and Peninsular sub-populations. Sci. Rep. 2023 doi: 10.1038/s41598-023-29368-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi R.K., Das M.K., Singh S.S., Sharma Y.D. Discordance in drug resistance-associated mutation patterns in marker genes of Plasmodium falciparum and Plasmodium knowlesi during coinfections. J. Antimicrob. Chemother. 2013;68:1081–1088. doi: 10.1093/jac/dks508. [DOI] [PubMed] [Google Scholar]
- Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic. Acids. Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Eede P., Vythilingam I., Ngo D.T., Nguyen V.H., Le X.H., D'Alessandro U., et al. Plasmodium knowlesi malaria in Vietnam: some clarifications. Malar. J. 2010;9(20) doi: 10.1186/1475-2875-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schalkwyk D.A., Blasco B., Davina Nunez R., Liew J.W.K., Amir A., Lau Y.L., et al. Plasmodium knowlesi exhibits distinct in vitro drug susceptibility profiles from those of Plasmodium falciparum. Int. J. Parasitol. Drugs Drug Resist. 2019;9:93–99. doi: 10.1016/j.ijpddr.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schalkwyk D.A., Moon R.W., Blasco B., Sutherland C.J. Comparison of the susceptibility of Plasmodium knowlesi and Plasmodium falciparum to antimalarial agents. J. Antimicrob. Chemother. 2017 doi: 10.1093/jac/dkx279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Lee Cs Fau - Bayoumi R., Bayoumi R Fau - Djimde A., Djimde A, Fau - Doumbo O., Doumbo O, Fau - Swedberg G., Swedberg G, Fau - Dao L.D., et al. Resistance to antifolates in Plasmodium falciparum monitored by sequence analysis of dihydropteroate synthetase and dihydrofolate reductase alleles in a large number of field samples of diverse origins. Mol. Biochem. Parasitol. 1997 doi: 10.1016/s0166-6851(97)00114-x. [DOI] [PubMed] [Google Scholar]
- White N.J. Plasmodium knowlesi: the fifth human malaria parasite. Clin. Infect. Dis. 2008;46:172–173. doi: 10.1086/524889. [DOI] [PubMed] [Google Scholar]
- WHO . 2021. World Malaria Report 2021. Retrieved from Geneva. [Google Scholar]
- WHO . 2022. WHO Guidelines for Malaria. 31 March 2022. Retrieved from Geneva. [Google Scholar]
- Yogavel M., Nettleship J.E., Sharma A., Harlos K., Jamwal A., Chaturvedi R., et al. Structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase-dihydropteroate synthase from Plasmodium vivax sheds light on drug resistance. J. Biol. Chem. 2018;293:14962–14972. doi: 10.1074/jbc.RA118.004558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaniyama J., Chitnumsub P., Kamchonwongpaisan S., Vanichtanankul J., Sirawaraporn W., Taylor P., et al. Insights into antifolate resistance from malarial DHFR-TS structures. Nat. Struct. Biol. 2003;10:357–365. doi: 10.1038/nsb921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.