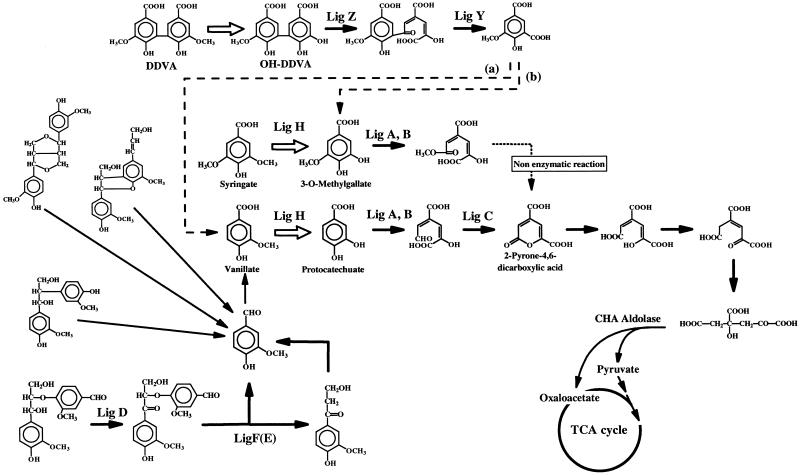
FIG. 1.
Degradation pathway of various lignin-related compounds in S. (Pseudomonas) paucimobilis SYK-6 (14). The O demethylation steps of phenylmethylether are represented by solid arrows. The enzymes catalyzing each step are indicated on the figure as follows: Lig A, B, protocatechuate 4,5-dioxygenase small and large subunits, respectively (27); Lig C, 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase (26); Lig D, arylglycerol-β-aryl ether Cα dehydrogenase (20); Lig F(E), α-keto-α-arylglycerol-β-aryl ether β-etherase isozymes (21, 22); Lig Z, OH-DDVA dioxygenase (11). OH-DDVA is converted to 5-carboxyvanillate with LigZ and LigY. Lig H, an enzyme related to the O demethylation of vanillate and syringate (this study). 5-Carboxyvanillate decarboxylase (represented by dotted arrow a) activity was detected in S. paucimobilis SYK-6 cell extract (24), and 3-O-methylgallate was detected by the metabolic experiment with 5-carboxyvanillate (represented by dotted arrow b) (14). The 2-pyrone-4,6-dicarboxylic acid metabolic pathway is represented according to findings from previous studies (18, 19, 26, 31). CHA aldolase, 4-carboxy-4-hydroxy-2-adipic acid aldolase; TCA cycle, tricarboxylic acid cycle.