Figure 2.
Harnessing natural transformation to generate mutant Saccharibacteria
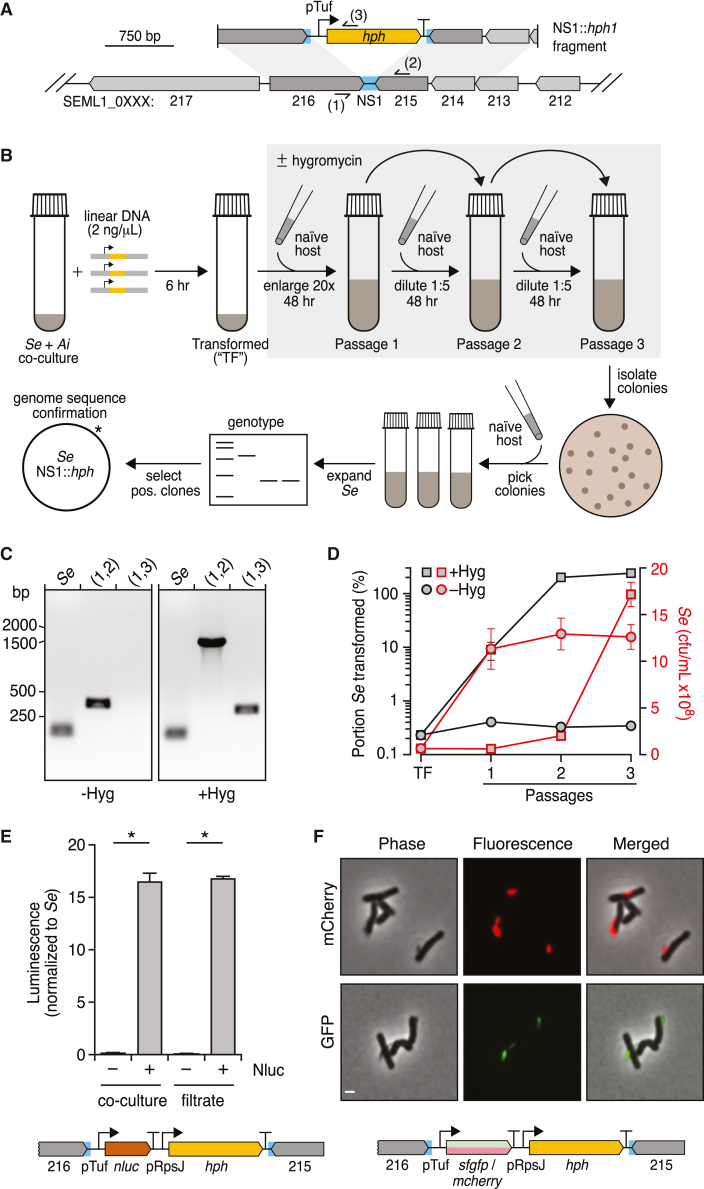
(A) Schematic depicting the intergenic neutral site (blue, NS1) targeted for insertion of a hygomycin resistance cassette (yellow) in the Se genome and the linear DNA fragment employed in transformation experiments. Primer binding sites used for genotyping are indicated (sites 1–3).
(B) Overview of the Se transformation protocol. After incubation with linear DNA, Se + Ai co-cultures are enlarged concomitant with hygromycin addition and serially passage with addition of naive host at each dilution to promote Se growth (gray box). Clonal transformed Se populations were obtained by plating to isolate single colonies of Ai with accompanying Se cells, followed by growth in liquid culture, with additional Ai, to promote Se population expansion. The asterisk indicates the single insertion detected by genome sequencing.
(C) PCR-based genotyping of Se clones obtained following transformation according to the protocol shown in (B) in the presence (right) or absence (left) of selection with hygromycin during the expansion and passaging steps. Binding sites for primers targeting NS1 (1, 2) and hph (3) are shown in (A). Positive control primers (Se) target a locus distant from NS1.
(D) Se growth (red) and percent of Se transformed (black) over the course of transformation protocol depicted in (B), in the presence (squares) or absence (circles) of selection with hygromycin.
(E) Luminescence production from Se-Ai co-cultures (left) or co-culture filtrates (right) in which Se contains a nanoluciferase expression cassette inserted at NS1 (shown at bottom).
(F) Fluorescence and phase contrast micrographs of Se-Ai co-cultures in which Se carries an mCherry (top) or sfgfp (bottom) expression cassette inserted at NS1. Scale bar, 1 μm. Data in (D) and (E) represent mean ± SD. Asterisks indicate statistically significant differences (unpaired two-tailed Student’s t test; ∗p < 0.05).
See also Figures S1 and S2.