Figure 3.
Fluorescent protein expression and quantitative microscopy enable tracking of the S. epibionticum lifecycle
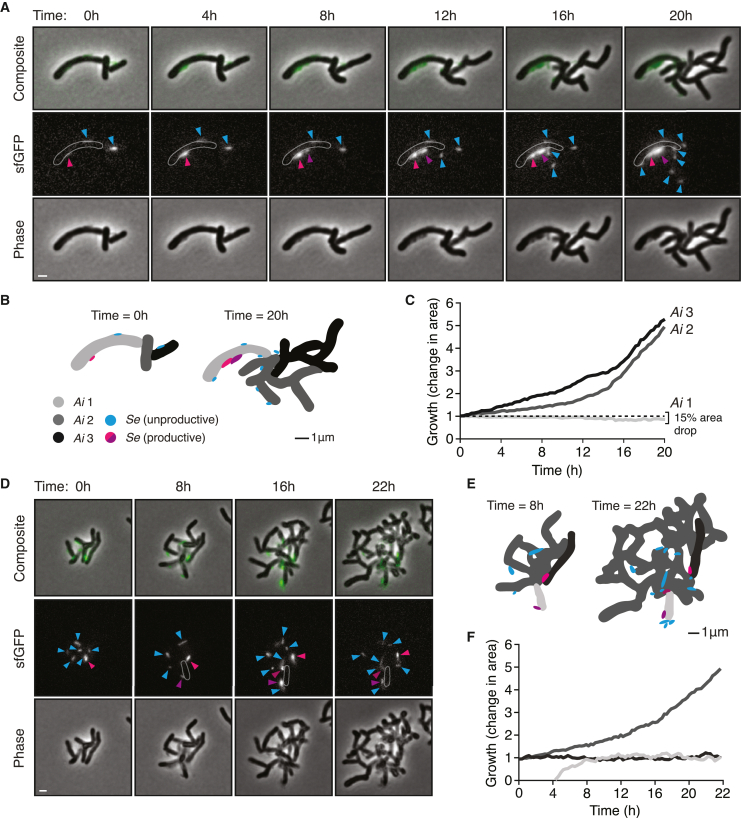
(A and D) Snapshots captured at the indicated time points from time-lapse fluorescence and phase contrast microscopy of GFP-expressing Se grown in co-culture with Ai. Arrows indicate example Se cells exhibiting productive (pink, purple) and non-productive (blue) interactions with Ai cells. White outlines in the fluorescent channel depict an Ai cell affected by Se infection (Ai 1). Scale bar, 1 μm.
(B and E) Omnipose-generated segmentation of Se and Ai cells depicted in (A) and (D), at the start (left) and end (right) of the 20- or 22-h growth period.
(C and F) Growth of individual Ai cells as impacted by productive (light gray) or non-productive Se cells (black, dark gray). Colors correspond to cell masks shown in (B) and (E). Time-lapse fluorescence microscopy videos from which (A) and (D) derive, as well as additional raw and annotated videos of Se-Ai co-cultures, are open for access via https://mougouslab.org/data.