Figure 4.
Identification of genes important for fitness of S. epibionticum during co-culture with Ai identified by Tn-seq
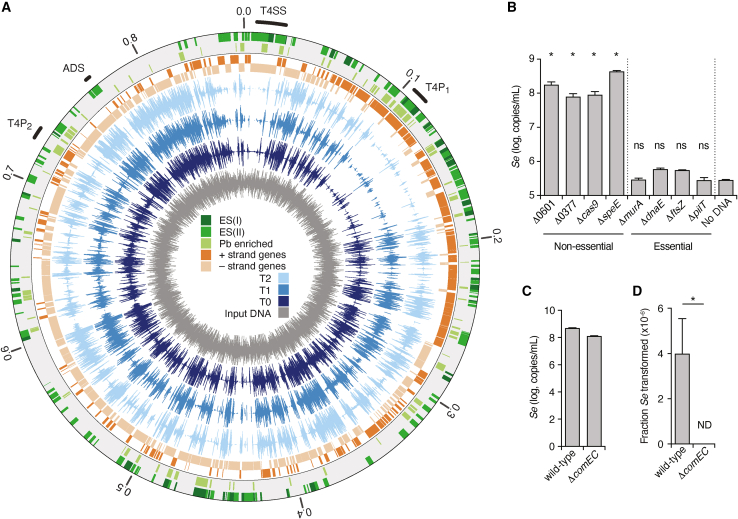
(A) Overview of normalized transposon-insertion frequency across the Se genome detected in input DNA used for mutagenesis (dark gray), and from samples collected 48 h after the onset of selection (T0, dark blue) and subsequent outgrowth time points (T1-T2, shades of blue). Genes encoding proteins belonging to Patescibacteria-enriched protein families (Pb enriched) and class I and II essential genes (ES I and II) are indicated in the outer circles (shades of green). The locations of the essential arginine deiminase system (ADS) genes, T4SS genes, and two loci containing T4P genes (T4P1 and T4P2) are indicated outside of the circle.
(B) Se population levels detected in Se-Ai co-cultures following transformation with constructs designed to replace the indicated genes with hph.
(C and D) Total Se population (C) and proportion transformed (D) following transformation with an unmarked cassette targeted to NS1 in the indicated strains of Se. See also Figures S4 and S5 and Table S1. Data in (B)–(D) represent mean ± SD. Asterisks indicate statistically significant differences (B, one-way ANOVA followed by Dunnett’s compared to no DNA control; D, unpaired two-tailed Student’s t test; ∗p < 0.05, ns, not significant).
See also Figures S3 and S4 and Table S1.