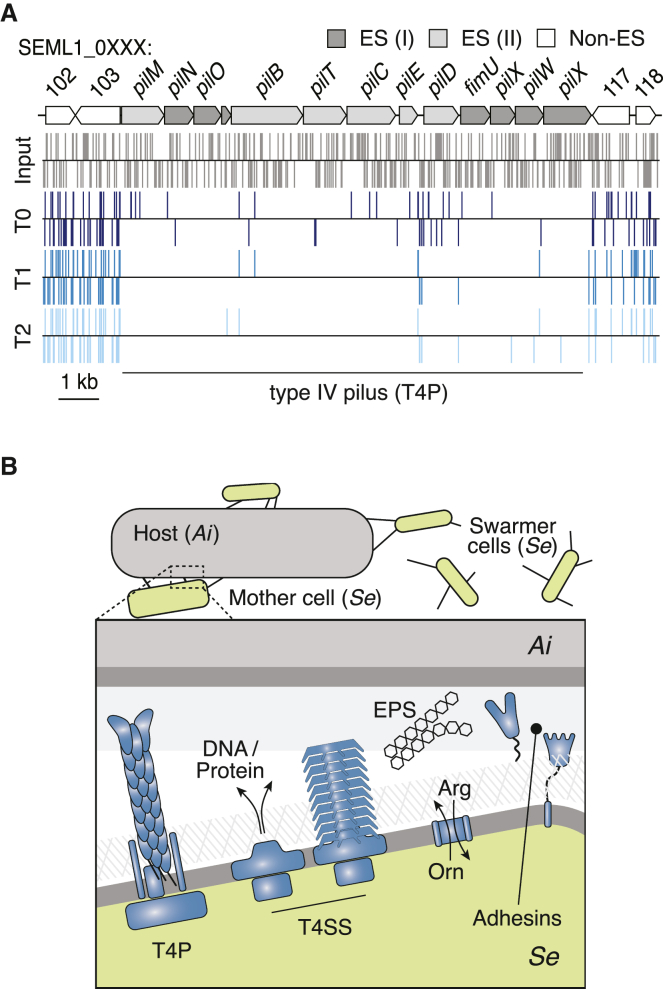
Figure 6.
Unusual essential genes of S. epibionticum encode numerous envelope-associated functions predicted to mediate host-cell interaction
(A) Schematic of genes encoding the core T4P components (T4P1 locus, top), and locations of transposon insertions detected in input mutagenized DNA (gray) and Tn-seq samples T0-T2 (shades of blue). Gene essentiality is indicated by shading (class I essential genes [ES I], dark gray; class II essential genes [ES II], light gray).
(B) Model of the Se/Ai interface during a productive infection, depicting selected envelope-associated functions found to be essential in our Tn-seq screen. These include macromolecular structures predicted to mediate adhesion (T4P) and DNA or protein delivery (T4SS), protein adhesins and the ADS system for arginine catabolism. Mother and swarmer cell designations derive from the analysis shown in Figure 3. See also Figure S5D. EPS, extracellular polysaccharide; Orn, ornithine; Arg, arginine.